Artificial Intelligence-Based Colorectal Polyp Histology Prediction by Using Narrow-Band Image-Magnifying Colonoscopy
Article information
Abstract
Background/Aims
We have been developing artificial intelligence based polyp histology prediction (AIPHP) method to classify Narrow Band Imaging (NBI) magnifying colonoscopy images to predict the hyperplastic or neoplastic histology of polyps. Our aim was to analyze the accuracy of AIPHP and narrow-band imaging international colorectal endoscopic (NICE) classification based histology predictions and also to compare the results of the two methods.
Methods
We studied 373 colorectal polyp samples taken by polypectomy from 279 patients. The documented NBI still images were analyzed by the AIPHP method and by the NICE classification parallel. The AIPHP software was created by machine learning method. The software measures five geometrical and color features on the endoscopic image.
Results
The accuracy of AIPHP was 86.6% (323/373) in total of polyps. We compared the AIPHP accuracy results for diminutive and non-diminutive polyps (82.1% vs. 92.2%; p=0.0032). The accuracy of the hyperplastic histology prediction was significantly better by NICE compared to AIPHP method both in the diminutive polyps (n=207) (95.2% vs. 82.1%) (p<0.001) and also in all evaluated polyps (n=373) (97.1% vs. 86.6%) (p<0.001)
Conclusions
Our artificial intelligence based polyp histology prediction software could predict histology with high accuracy only in the large size polyp subgroup.
INTRODUCTION
Colonoscopy with polypectomy of early colorectal neoplastic lesions (polyps) is a proven and widely accepted method of reducing colorectal cancer mortality rates. Predicting histology prior to endoscopic colorectal polyp removal is useful especially for diminutive (1–5 mm) and small (6–10 mm) polyps. Colorectal polyp histology can be non-neoplastic (hyperplastic) or neoplastic (tubular, villous adenomas, or sessile serrated lesions [SSLs]).
Non-neoplastic lesions, especially if they are diminutive hyperplastic polyps, may not require endoscopic polypectomy because of the negligible risk for developing malignancy [1-3].
Evaluation of colorectal polyps using the narrow-band imaging (NBI) technique and the NBI International Colorectal Endoscopic (NICE) classification are useful to predict the histology during endoscopy [3-7]. However, NBI and magnification based polyp histology prediction needs training and endoscopic experience. Moreover, the final and objective diagnosis still requires histology.
Therefore, we have been developing artificial intelligence-based polyp histology prediction (AIPHP) software to automatically evaluate the magnified NBI colonoscopy images aiming the histology prediction of polyps. We report the development of our software that can evaluate colorectal polyps using selectively recorded still colonoscopic images.
We aimed to analyze the AIPHP and NICE classification-predicted histology results and compare the histological predictive accuracy of the two methods.
MATERIALS AND METHODS
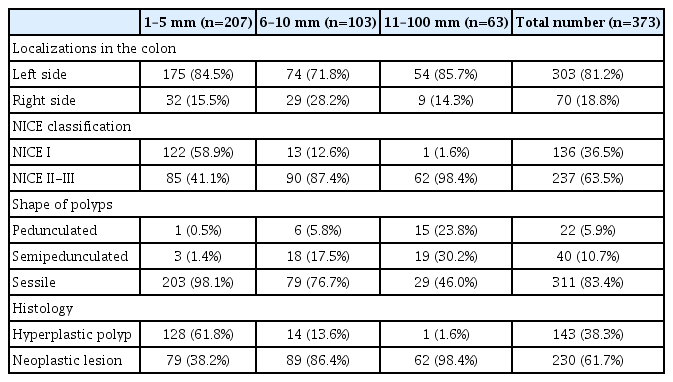
We examined 373 colorectal polyps (207 polyps, ≤5 mm; 103 polyps, 6–9 mm; and 63 polyps, ≥10 mm) obtained from 279 patients. Polyps were removed by traditional polypectomy or with mucosectomy or by cold-snare technique.
All endoscopic procedures and histological examinations were performed at the Petz Aladar University Teaching Hospital between October 2015 and November 2018.
Colonoscopies were performed with Olympus EXERA III CFHQ190 I (Olympus, Tokyo, Japan) high-resolution NBI colonoscope, providing 65x optical magnification.
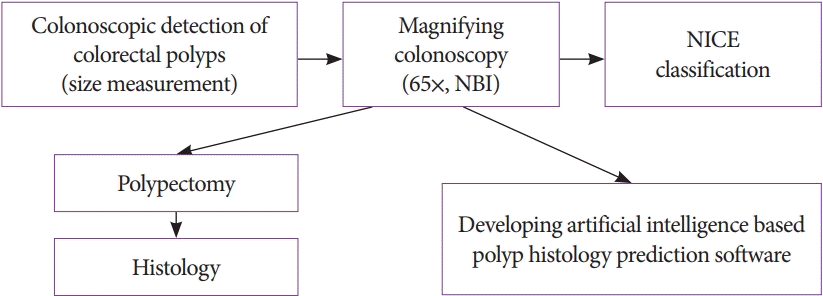
Colorectal polyps were detected first by high definition colonoscopy then by NBI at the optical maximum magnification (65×). All studied polyps were photo-documented. The stored NBI photos were analyzed by the NICE classification and AIPHP parallel system (Fig. 1).
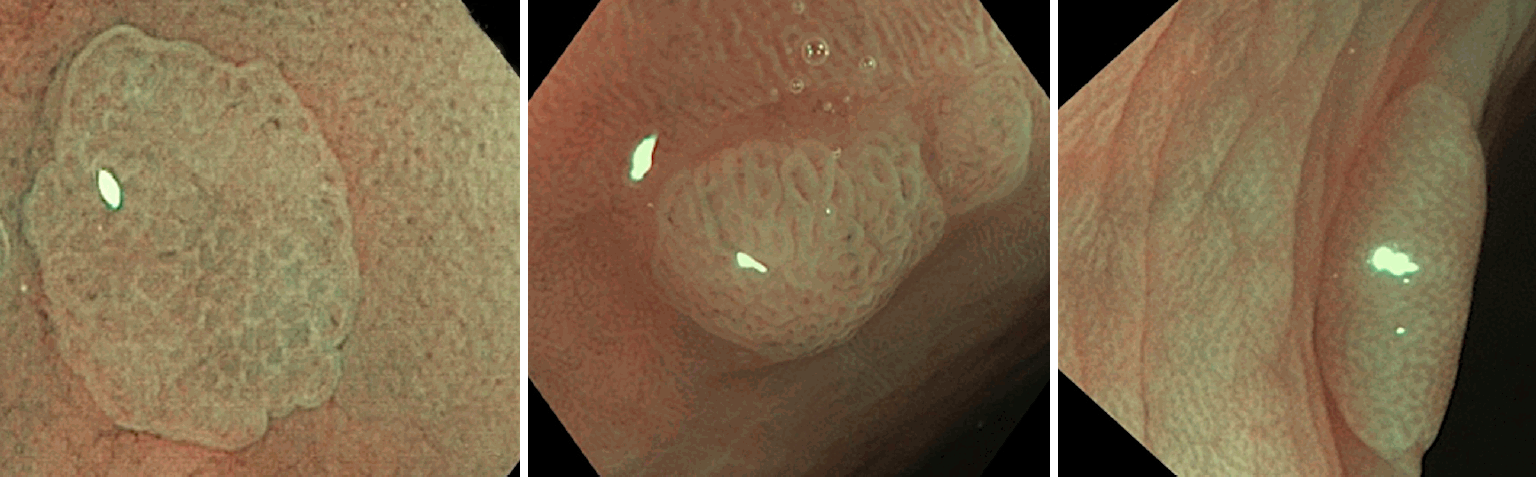
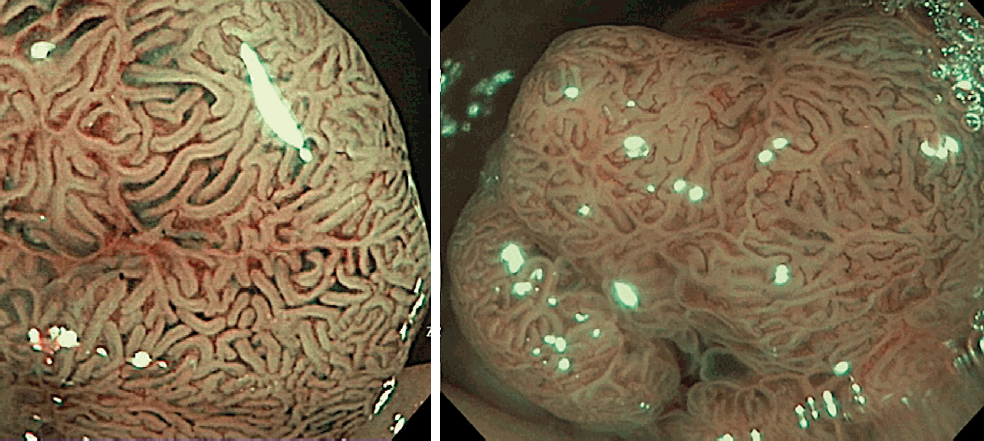
The still NBI-magnifying colorectal polyp images were taken before polyp removal by an endoscopist having >20-year experience. The stored NBI images were classified as type I or II-III with high-confidence prediction by three experienced endoscopists using NICE. All the three endoscopists were blinded to the histology and AIPHP results. In particular polyp cases when the NICE class assessment differed among the evaluating endoscopists, the majority (two-thirds) classification was accepted as final. The NICE classification divides pit patterns and microvessels at the surface structure in NBI images into types I, II, or III [4].
This classification correlates with the most likely pathology (Figs. 2, 3). NICE I corresponds to serrated lesions, such as hyperplastic polyps and SSLs, whereas NICE II and III correspond to adenomatous polyps.
Histological examination methods
Pathological examinations were performed by a single pathologist who was blinded to both the NICE and AIPHP system diagnosis. To determine the alterations, we used the WHO classification of colorectal polyps [8].
We used histology as the gold standard reference in statistical calculations. The two-class classifications were considered: hyperplastic or neoplastic (SSLs, tubular or villous adenomas, and invasive adenocarcinomas).
The study was approved by the Regional and Hospital Research Ethics Committee (ethical approval number: 76–1–20/2015.) and it was performed at the Department of Endoscopy and Gastroenterology, Petz Aladar University Teaching Hospital, Gyor, Hungary, in accordance with the Declaration of Helsinki (clinical trial registration number: NCT04425941). Patients gave informed consent for the endoscopic procedure and the pathological examination study as well as for the NICE and AIPHP analysis of the resected specimen.
AIPHP software system
The AIPHP software was developed in Python language, using OpenCV module. The AIPHP software is based on the categorization of the vascular pattern and color of the polyps.
The main steps of AIPHP software development were the following: 1) feature vector calculation, 2) training of classifier module, and 3) AIPHP classifier testing (Fig. 4). The present AIPHP version cannot automatically find the area of interest of polyp in a colonoscopy image; therefore human interaction is needed to select it. A simple image editor program (GNU Image Manipulation Program; GIMP, USA) was used for this purpose. The next step is the pre-processing that contains automatic noise reduction, glare removal, and a brightness correction step [9].

Main steps of artificial intelligence-based polyp histology prediction (AIPHP) training; feature vector calculation (left) and training of sub-classifiers (right). NBI, narrow-band imaging, HP, hyperplastic polyp; PI, polyp investigation; SSA, sessile serrated adenoma; SVM, support vector machine; TVA, tubulo villous adenoma.
Five features were used by our AIPHP software. Feature 1 is the relative standard deviance of the intensity diagram of the pre-processed image. Features 2 and 3 are the relative area of irregular bright and dark spots based on a method that classifies every spots as either “s1”, “s2”, “s3”, or “s4” where “s1” is for approximately circular and “s4” for irregular shapes with branches (Fig. 5). Features 4 and 5 measure the color difference of the polyp surface and the surrounding area.

Classification of bright and dark spots on a polyp surface. Blue, s1; green, s2; yellow, s3; red, s4.
Training was performed using a cross-validation scheme with 10 subsets. All trained 10 classification methods were saved and used later in the testing phase.
The average time to assess polyp histology by AIPHP was 0.5 ±0.2 s. A trained person had to mark the polyp surface on still images because the present AIPHP version cannot automatically find the area of interest. This maneuver took an additional 10–15 s (12.2±6).
Statistical analysis
Medical parameters and histological data were collected in one Excel file, which was analyzed by a self-developed Python program using the Pandas module to calculate various statistical quantities in tables. Fisher’s exact test was applied for calculating p-values using Scientific Python software (an open source, community developed software) and p<0.05 was considered statistically significant.
Linear regression was calculated to characterize the correlation between the size of polyps and accuracy of the AIPHP and NICE methods.
RESULTS
A total of 400 colorectal polyps were detected, photo-documented (NBI still images 65× magnification) and removed for histological analysis. Because of technical failures, like mechanical or thermal damage, 27 specimens were considered not suitable, hence a total of 373 polyps were characterized by AIPHP analysis and NICE classification as well as histologically (Fig. 6). The endoscopic, histological and NICE characteristics of the lesions are presented in Table 1.

Polyp analysis flowchart. AIPHP, artificial intelligence-based polyp histology prediction; NBI, narrow-band imaging; NICE, NBI international colorectal endoscopic; SD, standard deviation.
Among the polyps, 143 (38.3%) and 230 (61.7%) were hyperplastic and neoplastic (among them, 151 tubular adenomas, 70 tubulovillous adenomas, 3 SSLs and 6 invasive adenocarcinomas), respectively. Of the diminutive polyps, 128 (61.8%) and 79 (38.2%) showed hyperplastic and neoplastic histology, respectively (Table 1).
Utility of AIPHP and NICE classification
The accuracy of AIPHP was 86.6% (323/373) (sensitivity, 92.2%; specificity, 77.6%; positive predictive value [PPV], 86.9; negative predictive value [NPV], 86.0%) in all the polyps.
We compared the AIPHP accuracy results for diminutive and non-diminutive polyps (82.1% vs. 92.2%; p=0.0032), which showed a significantly higher accuracy in the non-diminutive polyp group. (Table 2).
Further, we evaluated the NICE classification results in predicting hyperplastic or neoplastic polyp histology in different polyp size groups. The accuracy of NICE results predicting the correct histology was 95.2% (197/207) (sensitivity, 97.5%; specificity, 93.7%; PPV, 90.6%; NPV, 98.4%) in the diminutive polyp group and 99.4% (165/166) (sensitivity, 100%; specificity, 93.3%; PPV, 99.3%; NPV, 100%) in the non-diminutive polyp group. (p=0.014) (Table 3)
The accuracy of the hyperplastic histology prediction was significantly better by NICE compared to AIPHP method both in the diminutive polyps (n=207, 95.2% vs. 82.1%, p<0.001) and also in all evaluated polyps (n=373, 97.1% vs. 86.6%, p<0.001) (Table 4)
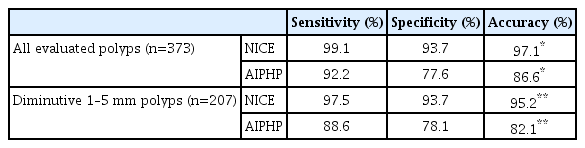
Hyperplastic Polyp Histology Predicting Data by NICE Classification and Artificial Intelligence-Based Polyp Histology Prediction
Neoplastic and hyperplastic histology were correctly predicted by AIPHP in 92.2% (212/230) and 77.6% (111/143) of the specimens, respectively (p<0.0001).
Polyp sizes had influenced both the AIPHP/histology and NICE/histology agreement results. A detailed calculation was performed to study the connection between the polyp sizes and agreement accuracy values using five size groups (Fig. 7).

Size dependency of NICE classification/histology and artificial intelligence-based polyp histology prediction (AIPHP)/histology agreement. r=0.568 for NICE classification/histology agreement (not significant), and r=0.918 for AIPHP/histology agreement (significant).
The accuracy of NICE prediction was close to 100% in all size groups whereas AIPHP/histology accuracy showed an increasing tendency with the polyp sizes. Linear regression correlation coefficient results indicate that AIPHP method was significantly more accurate in the bigger polyps than in the smaller polyps.
DISCUSSION
Colorectal polyp diagnostic methods have progressed very dynamically. The current dilemma is that diminutive polyps (≤5 mm) mostly show a hyperplastic histology which has a non-neoplastic behavior, and thus the rationale of polyp removal is under discussion.
High-definition endoscopy, including NBI technology, is able to predict the histology of colorectal polyps with high accuracy. However, this accuracy is significantly influenced by subjective elements, such as endoscopist technical skills and experience [6,10].
Consequently, the number of artificial intelligence polyp classification endoscopic methods is rapidly increasing [11-17].
The ideal aim of any AIPHP is to provide objective “semihistological” diagnosis and ensure high diagnostic accuracy for non-expert endoscopists.
In our study, the still images were taken by an endoscopist experienced with the NBI-based histology prediction criteria and able to choose good-quality images with optimal position. The photo-documented and stored polyp images were collected, and the regions of interest were selected manually.
We previously reported that our pilot AIPHP program can distinguish between images of hyperplastic or neoplastic lesions in NBI-magnifying colonoscopy images [18].
The Preservation and Incorporation of Valuable Endoscopic Innovation (PIVI) committee of the American Society for Gastrointestinal Endoscopy published a statement on establishing endoscopic diagnostic techniques [19-21]. Several NBI-based and magnifying endoscopy methods like NICE and Hiroshima classifications [22-25] are consistent with the PIVI recommendations of a ≥90% NPV and accuracy criteria.
Our present results show that the NICE classification fulfilled the PIVI criteria but the AIPHP program could only partly achieve PIVI recommendations among diminutive polyps (NPV, 91.7%; accuracy, 82.1%). However, in the case of non-diminutive polyps, the values fulfilled the PIVI criteria (sensitivity: 94.0%, accuracy: 92.2%). These findings indicate that the present AIPHP cannot offer the automatic computerized endoscopy-based histological diagnosis especially in those polyp size groups where the AI diagnosis would be most beneficial regarding cost-benefit considerations.
The question is why our AIPHP accuracy results, especially in the diminutive polyp subgroup, were worse than in the bigger polyp subgroups. One possible reason is that the diminutive polyps are mostly hyperplastic and are covered with mucous surfaces. The adherent mucous disturbs the ability to analyze the vascular pattern by AIPHP procedure. The mucous cover and the whitish shape is one reason why we got less precise histologically predictive results by AIPHP program compared to NICE classification results. Therefore, hyperplastic polyps can be differentiated better from adenomas using the NICE classification. Mucous surface and whitish characters hinder histological recognition by AIPHP program in which the polyp surface vascularity and the color differences are among the main features. In conclusion, we plan to add the mucus surface as a new feature for the next-generation AIPHP programs.
To the best of our knowledge, this is one of the first studies which analyzed the AI diagnostic results in different polyp sizes. We found that the AIPHP method is significantly more accurate in the bigger size polyps than in smaller ones. Polyp sizes had significantly influenced AIPHP accuracy but not the NICE/histology agreement results. This study is also pioneering in comparing the NICE classification with a special artificial intelligence-based polyp image analyzer. Both hyperplastic and SSLs correspond to NICE I, therefore it is almost impossible to distinguish the two by NICE classification. This problem limits the accuracy of NICE classification regarding the differentiation of non-neoplastic polyps and the neoplastic SSLs. In our study, the accuracy of NICE evaluation is high, which is partly due to the small number (n=3) of SSLs analyzed in the study. The accuracy of NICE likely would decrease as the ratio of SSLs increases.
We comparatively analyzed our AIPHP results with those three studies in which software-based automatic polyp histology analysis provided higher sensitivity and accuracy results than our outcomes. Takemura et al. [26] and by Komiani et al. [27] studied low numbers of hyperplastic polyps (47 and 45, respectively) which are notably less than the 143 hyperplastic polyps in our study. The critical point of the artificial intelligence (AI) analysis is to correctly identify the diminutive non-neoplastic lesions. To fulfill this requirement, an eligible number of diminutive hyperplastic lesions should be analyzed. In the study of Gross et al. [12], 135 non-neoplastic diminutive polyps were analyzed by a computer with 94.3% NPV. These results are slightly superior to our findings showing a 91.7% NPV by AIPHP in 128 hyperplastic lesions.
There are several limitations of our study. Trainings are necessary both for NICE evaluation and production of high-quality stored polyp images. Diminutive polyps are typically hyperplastic with a mucous covering, interfering both photo-documentation quality and correct AIPHP recognition. The “one polyp, one picture” method for analysis was suitable for NICE evaluation but did not complete the correct AI software analysis requirements in diminutive and small polyps. The present AIPHP version cannot automatically find the area of interest on the polyp surface, hence additional human interaction is needed. We found that 10–15 s is sufficient for a trained person to mark the polyp surface on a still image. It could also be acceptable in routine endoscopy, provided that the selection marker could be integrated in the software. Such an integrated software is under development.
Further problems that may limit the generalizability of our findings are that there are many less experienced endoscopists in the real-world practice who may not take good-quality images on particular areas of interest. In addition, magnifying endoscopy is not widely used in general endoscopy.
In conclusion, our artificial intelligence system manipulating still images of colorectal polyps by NBI-magnifying colonoscopy could predict histology with high accuracy only for non-diminutive (>5 mm) polyps. The AIPHP accuracy significantly increased with larger polyp sizes. Similar to other studies, NICE classification results showed high sensitivity and accuracy in all polyp size groups.
Because our present AIPHP cannot substitute the subjective endoscopic virtual histology evaluations, especially in diminutive polyps, further development with automatic area-of-interest detection and parallel analysis of multiple images of the same polyp could assist automatic polyp histology predictions, even in smaller colorectal polyps.
Notes
Conflicts of Interest: The authors have no potential conflicts of interest.
Funding:This study was supported by the GINOP-2.3.4-15-2016-00003 grant.
Author Contributions
Conceptualization: Istvan Racz, Zoltan Horvath
Data curation: Henriett Regoczi, Noemi Kranitz, Andras Horvath
Formal analysis: IR, AH
Funding acquisition: ZH
Investigation: IR, NK, Gyongyi Kiss
Methodology: IR, ZH
Project administration: HR
Resources: ZH
Software: AH, ZH
Supervision: IR, ZH
Validation: IR
Visualization: IR, GK, HR
Writing-original draft: IR
Writing-review&editing: IR, HR