AbstractBackground/AimsPrevious artificial intelligence (AI) models attempting to segment gastric intestinal metaplasia (GIM) areas have failed to be deployed in real-time endoscopy due to their slow inference speeds. Here, we propose a new GIM segmentation AI model with inference speeds faster than 25 frames per second that maintains a high level of accuracy.
MethodsInvestigators from Chulalongkorn University obtained 802 histological-proven GIM images for AI model training. Four strategies were proposed to improve the model accuracy. First, transfer learning was employed to the public colon datasets. Second, an image preprocessing technique contrast-limited adaptive histogram equalization was employed to produce clearer GIM areas. Third, data augmentation was applied for a more robust model. Lastly, the bilateral segmentation network model was applied to segment GIM areas in real time. The results were analyzed using different validity values.
ResultsFrom the internal test, our AI model achieved an inference speed of 31.53 frames per second. GIM detection showed sensitivity, specificity, positive predictive, negative predictive, accuracy, and mean intersection over union in GIM segmentation values of 93%, 80%, 82%, 92%, 87%, and 57%, respectively.
INTRODUCTIONGastric intestinal metaplasia (GIM) is a well-known premalignant lesion and a risk factor for gastric cancer.1 Its diagnosis is highly challenging due to subtle mucosal changes that can be easily overlooked. White light endoscopy (WLE) alone, when observed by less experienced endoscopists, may lead to premalignant lesions from normal mucosa being missed.2,3 Various techniques have been developed to enhance the detection rate of these lesions, including the multiple random biopsy protocol (Sydney protocol)4 and image-enhanced endoscopy (IEE), including narrow-band imaging (NBI), blue light imaging, linked color imaging, iScan, and confocal laser imaging.5-7 Random biopsy for histological evaluation is generally cost prohibitive. IEE diagnosis entails a significant time requirement for training endoscopists to achieve high accuracy in GIM interpretation. Unlike colonic polyps, segmentation of GIM is difficult to define because it usually has an irregular border with many scattered lesions. Segmentation of GIM can become very difficult and often results in poor results when performed by less experienced endoscopists.8
Deep learning models (DLMs) have made dramatic entrances into the field of medicine. One of the most popular research objectives in upper gastrointestinal (GI) endoscopy is to detect and segment early gastric neoplasms.9,10 Among the publicly available models, DeepLabV3+11 and U-Net12 are considered state-of-the-art DLMs for segmentation tasks. However, these models cannot be integrated in a real-time system that requires at least 25 frames per second (FPS)13; this is because of the significant computation requirements for high-resolution images, which result in delays and sluggish displays of the captured areas during real-time endoscopy.
Rodriguez-Diaz et al.14 applied DeepLabV3+ to detect colonic polyps and achieve an inference speed (IFS) of only 10 FPS. Wang et al.15 applied DeepLabV3+ to segment gastrointestinal metaplasia and found that the IFS was only 12 FPS. In addition, Sun et al.16 employed U-Net to detect colonic polyps and reached a speed close to the threshold IFS (22 FPS). However, the image size was still relatively small (384├Ś288 pixels) when compared with the much larger image in the current standard practice (1,920├Ś1,080 pixels). Findings from our earlier experiments with U-Net revealed that the IFS was only three FPS in the current standard image size. In addition, the detection performances of many DLMs are still limited owing to the availability of only a handful of training GIM datasets. The suboptimal supply of medical images might cause low accuracy in the model, leading to poor practice performance. Therefore, additional techniques to improve DLM accuracy, including transfer learning (TL),17 image enhancement, and augmentation (AUG), may be necessary.
This study aimed to establish and implement a new DLM with additional techniques (TL, image enhancement, and AUG) that could produce a more practical real-time semantic segmentation with high accuracy to detect GIM during upper GI endoscopy.
METHODSStudy design and participantsA single-center prospective diagnostic study was performed. We trained and tested our DLMs to detect GIM using WLE and NBI using data from the Center of Excellence for Innovation and Endoscopy in Gastrointestinal Oncology, Chulalongkorn University, Thailand. Informed consent for the endoscopic images was obtained from consecutive GIM patients aged 18 years or older who underwent upper endoscopy under WLE and/or IEE between January 2016 and December 2020. We assigned the pathological diagnosis as a ground-truth diagnosis. Two pathologists from King Chulalongkorn Memorial Hospital made pathological assessments of specimens obtained from different biopsy stomach sites in at least five areas: two antrums, two bodies, and one incisura, according to the updated Sydney System. Patients with a history of gastric surgery or otherwise diagnosed with other gastric abnormalities such as erosive gastritis, gastric ulcers, gastric cancer, high-grade dysplasia, low-grade dysplasia, and those without a confirmed pathological diagnosis were excluded.
Endoscopy and image quality controlAll images were recorded using an Olympus EVIS EXERA III GIF-HQ190 gastroscope (Olympus Medical Systems Corp., Tokyo, Japan). Two expert endoscopists (RP and KT) with a minimum of 5 years of experience in gastroscopy and a minimum of 3 years of experience in IEE and had performed more than 200 GIM diagnoses were selected to review all images. Poor-quality images, which consisted of halation, blur, defocus, and mucus, were removed. The size of the raw images was 1,920├Ś1,080 pixels, and they were stored in a joint photographic expert group (JPEG) format. All images were cropped to show only the gastric epithelium (nonendoscopic images and other labels such as patient information were removed), resulting in an image with 1,350├Ś1,080 pixels.
Image datasetsAfter the biopsy-proven GIM images were obtained, with unanimous agreement, two expert endoscopists (RP and KT) performed annotations on the images to define GIM segments using LabelMe.18 The labeled images were stored in portable network graphics (PNG) format and referred to as ground-truth images. Data were stratified by the type of image to ensure that the proportion between white light and NBI images was maintained. The labeled GIM images were separated into three datasets. Seventy percent of the expected total GIM images were used as the training set, 10% were assigned for validation, and 20% served as the test dataset.
In addition to GIM images, an equal number of non-GIM gastroscopy images were also included in the test set to represent a realistic situation and to evaluate the results on non-GIM images. In these non-GIM frames, no annotations were made, except for cropping of the nonendoscopic areas.
Model developmentFour main modules, including three preprocessing modules and one model training module, were used to improve the accuracy of our DLM (Fig. 1). First, the concept of TL was employed to overcome the small training size issue of GIM.17 We utilized 1,068 colonic-polyp images from public datasets (CVC-Clinic19 and Kvasir-SEG20), as shown in Supplementary Figure 1, to pretrain the model to allow the DLM to learn from the GIM dataset more effectively (Supplementary Table 1).
Second, and similar to IEE, each GIM image was enhanced by using contrast-limited adaptive histogram equalization (CLAHE) to amplify the contrast of the GIM regions (Supplementary Fig. 2).21 Third, multiple data AUG tricks were applied to create data variations. Nine AUG tricks were employed, including flips (horizontal and vertical), rotations (0┬░ŌĆō20┬░), sharpening, adding of noise, transposition, shift scale rotations, blur, optical distortions, and grid distortions. This step was aimed at increasing the training size, preventing overfitting, and making the model more robust (Supplementary Fig. 3).
Finally, a bilateral segmentation network (BiSeNet)22 specifically designed for real-time segmentation using a much smaller model was applied on top of the pretrained model containing both the enhanced and augmented GIM images to achieve an IFS greater than 25 FPS.13 Two baseline models (DeepLabV3+11 and U-Net12) were also used to analyze the datasets and serve as benchmark models (Supplementary Fig. 4).
Performance evaluationThe modelŌĆÖs performance was evaluated by an internal test in regards to two aspects: (1) GIM detection (the whole frame) and (2) GIM area segmentation (labeling the gastric mucosa area containing GIM). For GIM detection, true positives were classified when the prediction and the ground-truth regions in the GIM images overlapped by greater than 30%. True negatives were assigned when the size of the predicted region covered less than 1% of the non-GIM areas. The model validity was analyzed using metrics including sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy.
Segmentation performance was evaluated by the mean intersection over union (mIoU) areas, as shown in Figure 2. For non-GIM images, we calculated the surplus area that was incorrectly predicted as the GIM area. This was defined as the ŌĆ£errors.ŌĆØ IFS, a crucial measure that must be greater than 25 FPS in order for the model to create inferences in real time, was evaluated using FPS. The test dataset was assessed using each model. BiSeNet22 was used as the main model, with additional TL,17 CLAHE,21 and AUG techniques. We used four versions of our new models (BiSeNet alone, BiSeNet+TL, BiSeNet+TL+CLAHE, and BiSeNet+TL+CLAHE+AUG) in the test dataset. Two benchmark models (DeepLabV3+11 and U-Net12) were also tested on the same dataset to compare the model performance (Fig. 3).
Statistical analysisFor the classification result, McNemar test was conducted to compare agreement and disagreement between our model and each baseline. For the segmentation results, a paired t-test was conducted for each image. For the IFS, a paired t-test was also computed to compare the run time of each round (five rounds total).
Ethical statementsThe Institutional Review Board of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand has approved this study in compliance with the International Guidelines for Human Research Protection in the Declaration of Helsinki, the Belmont Report, CIOMS guidelines, and the International Conference on Harmonization in Good Clinic Practice (ICH-GCP; COA 1549/2020; IRB number 762/62). The protocol was registered at ClinicalTrials.gov (NCT04358198).
RESULTSWe collected and labeled 802 biopsy-proven GIM images from 136 patients between January 2016 and December 2020 from the Center of Excellence for Innovation and Endoscopy in Gastrointestinal Oncology, Chulalongkorn University, Thailand. A total of 318 images were obtained from WLE, and 484 were NBI. Two expert endoscopists (RP and KT) performed annotations on the images to define GIM segments using labeling software, LabelMe,18 with unanimous agreement. Labeled GIM images were randomly separated into training (70%, 560 images), validation (10%, 82 images), and testing (20%, 160 images) datasets (Table 1). The test dataset also included 160 non-GIM gastroscopy images, which included 137 WLE and 23 NBI images.
GIM diagnostic performanceUsing the images from both WLE and NBI, BiSeNet combined with three additional preprocessing techniques (TL, CLAHE, and AUG) showed the highest sensitivity (93.13%) and NPV (92.09%) when compared to BiSeNet alone and BiSeNet without full preprocessing techniques. The diagnostic specificity, accuracy, and PPV of BiSeNet+TL+CLAHE+AUG were 80.0%, 86.5%, and 82.3%, respectively. The overall performance of our proposed model (BiSeNet+TL+CLAHE+AUG) was significantly better than those of DeepLabV3+ and U-Net (p<0.01 for all parameters). The results for all six models are presented in Table 2.
The diagnostic performances of BiSeNet+TL+CLAHE+AUG when using WLE images alone (Table 3) was lower than that of NBI images alone (Table 4) in all modalities, including specificity (78.8% vs. 86.9%), accuracy (80.8% vs. 95.2%), and PPV (62.3% and 97.1%). Furthermore, the overall performance of BiSeNet+TL+CLAHE+AUG from either WLE or NBI images was significantly better than the benchmarks DeepLabV3+ and U-Net (p<0.01 for all parameters) (Tables 3, 4).
GIM segmentation performanceBiSeNet with three preprocessing models showed the highest mIoU in segmenting GIM areas (57.04%┬▒2.75%) compared to BiSeNet alone and BiSeNet without full preprocessing techniques (mIoU 45.94%┬▒3.07% for BiSeNet alone; 47.29%┬▒3.18% for BiSeNet+TL; and 54.94%┬▒2.90% for BiSeNet+TL+CLAHE), with an error of less than 1% (0.96%). When compared to the benchmark models DeepLabV3+ and U-Net, the mIoU of BiSeNet with the three combinations was significantly better (mIoU 57.04%┬▒2.75% for BiSeNet+TL+CLAHE+AUG; 49.22%┬▒3.06% for DeepLabV3+; and 53.02%┬▒2.99% for U-Net; p<0.01 in all parameters) (Table 5).
The segmentation performance of BiSeNet+TL+CLAHE+AUG when using WLE images alone (Supplementary Table 2) was lower than that using NBI images alone (Supplementary Table 3), 52.94% vs. 59.25% in terms of mIoU. Moreover, the overall performance of BiSeNet+TL+CLAHE+AUG using either WLE or NBI images was significantly better than that of DeepLabV3+ and U-Net (p<0.01) (Supplementary Tables 2, 3).
Inference speedThe IFS of BiSeNet+TL+CLAHE+AUG was 31.53┬▒0.10 FPS. BiSeNet alone and all BiSeNet combinations with preprocessing models achieved an IFS greater than the 25 FPS threshold. The IFS of the benchmark models, DeepLabV3+ and U-Net, reached only 2.20┬▒0.01 and 3.49┬▒0.04, respectively (Table 6).
To explore the ability of our model to segment GIM areas in a real-time clinical setting, we tested the model on a WLE video (Supplementary Video 1). In the video clip, the model successfully segmented the GIM lesions correctly without any sluggishness.
DISCUSSIONThe suspicion of GIM based on artificial intelligence (AI) readings may facilitate targeted biopsy. In particular, since unnecessary endoscopic biopsy can be avoided, AI methods would be useful for patients at risk of bleeding, such as those with coagulopathy or platelet dysfunction or those taking antithrombotic agents. Techniques using deep learning for the detection and analysis GI lesions using convolutional neural networks23 have rapidly evolved.24 The two primary objectives for DLM are detection and diagnosis (CADe/CADx). Earlier DLMs for GI endoscopy have been successfully employed on the lower GI tract (e.g., colonic-polyp classification, localization, and detection). DLM employed on the colon achieved a real-time performance with a very high sensitivity (>90%).25
There have been many attempts to use DLM to detect upper GI lesions, including gastric neoplasms, while performing upper GI endoscopy; however, most studies have focused on detecting gastric cancer.26-29 Unlike colonic polyps that require only CADe/CADx, GIM requires more precise segmentation because it usually contains irregular borders with many satellite lesions. None of the current DLMs could achieve the expected level of real-time segmentation because the IFS was still too slow. Xu et al.30 used four DLMs, including ResNet-50,31 VGG-16,32 DenseNet-169,33 and EfficientNet-B4,34 and demonstrated their GIM detection capabilities; however, the endoscopist still had to freeze the image in order to segment the GIM area. This is not practical when performing a real-time upper GI endoscopy.30 In addition, their DLMs were mainly used for interpreting NBI images rather than WLE images, which is usually the preferred mode during initial endoscopy.
Our study showed that by adding all three preprocessing techniques (TL, CLAHE, and AUG) to the BiSeNet model, the new DLM could achieve a sensitivity and NPV higher than 90% for detecting GIM using both WLE and NBI images while maintaining an IFS faster than the 25 FPS threshold.
BiSeNet is a recent real-time semantic segmentation tool that balances the need for accuracy with an optimum IFS. Using BiSeNet alone, the IFS was ten times faster than the two other baseline models, DeepLabV3+ and U-Net (34.02 vs. 2.20 and 3.49% FPS), with comparable performance in terms of classification and segmentation. Despite the impressive speed, the sensitivity and NPV of BiSeNet alone for GIM detection was only 82% and 83%, respectively. Hence, it could not pass the threshold recommended by the Preservation and Incorporation of Valuable Endoscopic Innovations (PIVI) standards for diagnostic tools that require an NPV>90%.35 Therefore, additional preprocessing methods such as TL, CLAHE, and AUG are required to improve model validity, especially the NPV.
While the number of GIM images in our database was limited, the upper GI images shared common characteristics with colonoscopic images in terms of color and texture, allowing the colonoscopic images to be utilized for TL. By adding 2,680 colonoscopic images, TL increased the specificity of the model from 87% to 92%, although the NPV remained lower than the 90% threshold.
The biggest improvement in our DLM can be credited to the application of CLAHE. The detection sensitivity improved by almost 10% (from 82% to 89%), and the mIoU increased by 9% points (from 46% to 55%). This is probably due to the large number of WLE images in our training data, which provided robust image enhancement. Because CLAHE enhances small details, especially textures and local contrast, it can amplify WLE image contrast similar to IEE technology. This is promising for achieving high efficacy in our DLM on WLE images without the need for the NBI mode during real-time endoscopy.
AUG also further improved the sensitivity compared to the previous model by 4% (from 89% to 93%) and increased the mIoU by 2% (from 55% to 57%). Among all three preprocessing methods, TL seemed to show the least benefit. We believe that this may be caused by the differences in the backgrounds of the pretraining colon datasets when compared to the GIM dataset. In retrospect, other upper GI images of disorders such as hemorrhagic gastritis, gastric ulcer, and gastric cancer should have been used instead of just colon images.
Our study illustrated that when using BiSeNet alone, the IFS (31.53┬▒0.10) was faster than the minimum requirement for a real-time performance of 25 FPS. Although the model might have a comparable sensitivity to the two original benchmarks, DeepLabV3+ and U-Net (81.88% vs. 83.75% and 87.50%, respectively), the specificity of BiSeNet was significantly higher than that of the two baselines (87.50% vs. 70.00% and 62.50%, respectively; p<0.01). By adding the three techniques of TL, CLAHE, and AUG, we demonstrated a significant improvement in validities across the board. Importantly, the high NPV for GIM diagnosis in our model (92.09%) exceeded the acceptable performance threshold outlined by PIVI as a screening endoscopy tool. Notably, the other DLMs did not reach the threshold number.
For the GIM segmentation, our DLM produced an mIoU of 57.04%, which is considerably lower than that of a substantial mIoU. Since GIM typically presents as scattered lesions in the same area, we believe that correct segmentation on more than half of all GIM lesions is sufficient for the endoscopists to perform targeted biopsy and to provide the correct recommendation regarding the frequency of endoscopic surveillance from the extension of GIM. For example, the British Society of Gastroenterology guidelines on the diagnosis and management of patients at risk of gastric adenocarcinoma recommend an interval of endoscopic surveillance according to the extension of GIM.36 An interval of 3 years is recommended for patients with extensive GIM, defined as that affecting both the antrum and body. For patients with GIM limited to only the antrum, the risk of gastric cancer development is very low; therefore, further surveillance is not recommended.36
The key breakthrough of this study is that, for the first time, a DLM has been able to achieve an IFS speed that is fast enough to perform real-time GIM segmentation. Our full DLM could also detect and segment GIM areas from both WLE and NBI images. We believe that our DLM is more practical for endoscopists who usually perform initial endoscopy with WLE and then switch to NBI mode for detailed characterization after a lesion is detected. We also feel that our DLM may aid less experienced endoscopists in locating more suspected GIM areas during WLE, considering that they can switch to NBI mode after being notified by the DLM to further examine the suspected GIM areas more efficiently.
The findings of this study must be tempered with several limitations that could impact the successful replication and real-time success of our model. First, we retrieved the GIM images from one endoscopy center using a single endoscope model. To address the issue of dataset quality and increase the generalizability of results, images from other endoscopy centers and endoscope models with different IEE modes, including iScan, blue light imaging, and linked color imaging, are needed. Second, this model has not been fully studied in a real-time setting. Our preliminary test showed that it could function well without sluggish frames. Most importantly, endoscopists did not need to freeze the video to produce the still image for the DLM analysis (Supplementary Video 1). Third, we excluded other endoscopic findings, including hemorrhagic gastritis, gastric ulcer, and gastric cancer, as well as retained food content, bubbles, and mucous from our datasets. Therefore, our model may return more errors when analyzing lesion images. However, we feel that in real-time procedures, endoscopists should easily distinguish between these abnormalities and the actual suspected GIM areas. Finally, the results of this study were based on an internal test that lacked external validation. Nevertheless, we plan to conduct an external validation test to prove the results of this study in the near future.
In conclusion, compared with the benchmark models DeeplabV3+ and U-Net, the BiSeNet model in combination with three techniques (TL, CLAHE, and data AUG) significantly improved GIM detection while maintaining a fair quality of segmentation (mIoU>50%). With the IFS reaching 31.53 FPS in this model, these results pave the way for future research on real-time GIM detection during upper GI endoscopy.
Supplementary MaterialSupplementary Video 1. The model was tested on a real video during an esophagogastroduodenoscopy. It successfully captured the gastric intestinal metaplasia lesions in real time (https://doi.org/10.5946/ce.2022.005.v001). Supplementary Fig. 3. Examples of the nine data augmentation techniques applied to each gastric intestinal metaplasia image. Supplementary Table 1. Images and resolution of each dataset for transfer learning. Supplementary Table 2. Segmentation performance of two baseline models (DeepLabV3+ and U-Net) compared to four BiSeNet family models focused on white light endoscopy images. Supplementary Table 3. Segmentation performance of two baseline models (DeepLabV3+ and U-Net) compared to four BiSeNet family models focused on narrow-band imaging images. Supplementary materials related to this article can be found online at https://doi.org/10.5946/ce.2022.005.
NOTESConflicts of Interest
The GPUs used in this project were sponsored by an NVIDIA GPU grant in collaboration with Dr. Ettikan K Karuppiah, a director/technologist at NVIDIA, Asia Pacific South Regions. The authors have no other conflicts of interest to declare.
Funding
This research was funded by the National Research Council of Thailand (NRCT; N42A640330), Chulalongkorn University (CU-GRS-64), and Chulalongkorn University (CU-GRS-62-02-30-01) and supported by the Center of Excellence in Gastrointestinal Oncology, Chulalongkorn University annual grant. It was also funded by the University Technology Center (UTC) at Chulalongkorn University.
Author Contributions
Conceptualization: RP, PV, RR; Data curation: VS, RP, KT, NF, AS, NK; Formal analysis: VS, RP, KT, PV, RR; Funding acquisition: PV, RR; Methodology: RP, PV, RR; WritingŌĆōoriginal draft: VS, KT; WritingŌĆōreview & editing: RP, PV, RR. All authors read and approved the final manuscript.
Fig.┬Ā1.The proposed framework of our study. GI, gastrointestinal; CLAHE, contrast-limited adaptive histogram equalization; BiSeNet, bilateral segmentation network. 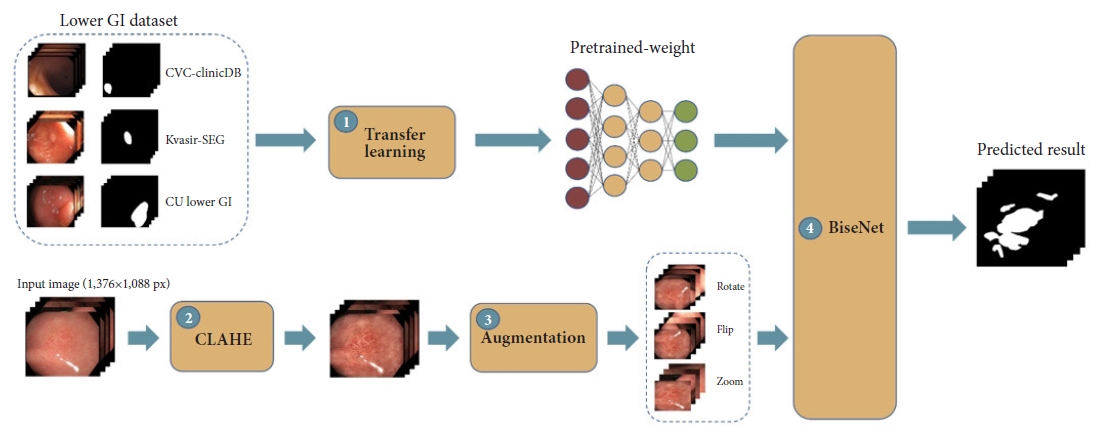
Fig.┬Ā2.Examples of intersection over union (mIoU) evaluation on a gastric intestinal metaplasia image. (A) IoU=0.8, (B) IoU=0.6, (C) IoU=0.4. Red indicates a ground-truth region. Blue indicates a predicted region. Green demonstrates the intersected area. 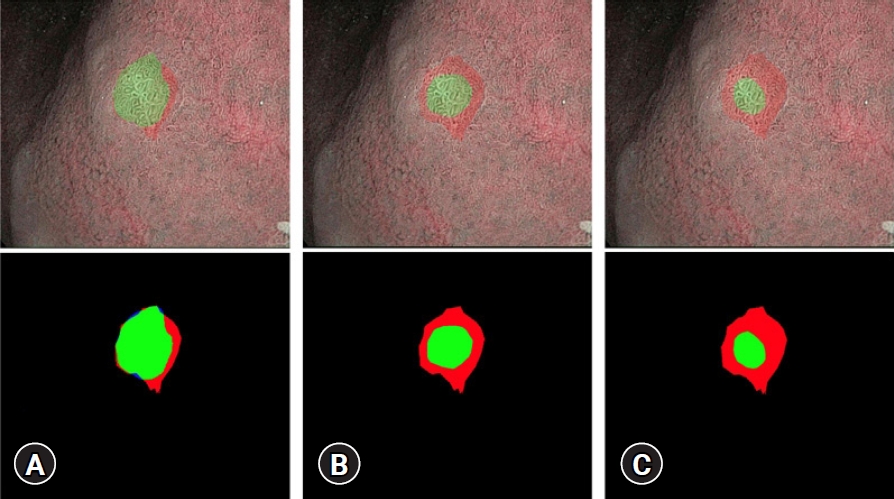
Fig.┬Ā3.Prediction examples in six images, where the green circle encloses the gastric intestinal metaplasia (GIM) area. (A) Raw image, (B) ground-truth, and (C) prediction by BiSeNet alone, and (D) prediction by our full model (BiSeNet+TL+CLAHE+AUG). Rows 1ŌĆō4 represent GIM images, and rows 5ŌĆō6 represent non-GIM images. BiSeNet, bilateral segmentation network; TL, transfer learning; CLAHE, contrast- limited adaptive histogram equalization; AUG, augmentation. 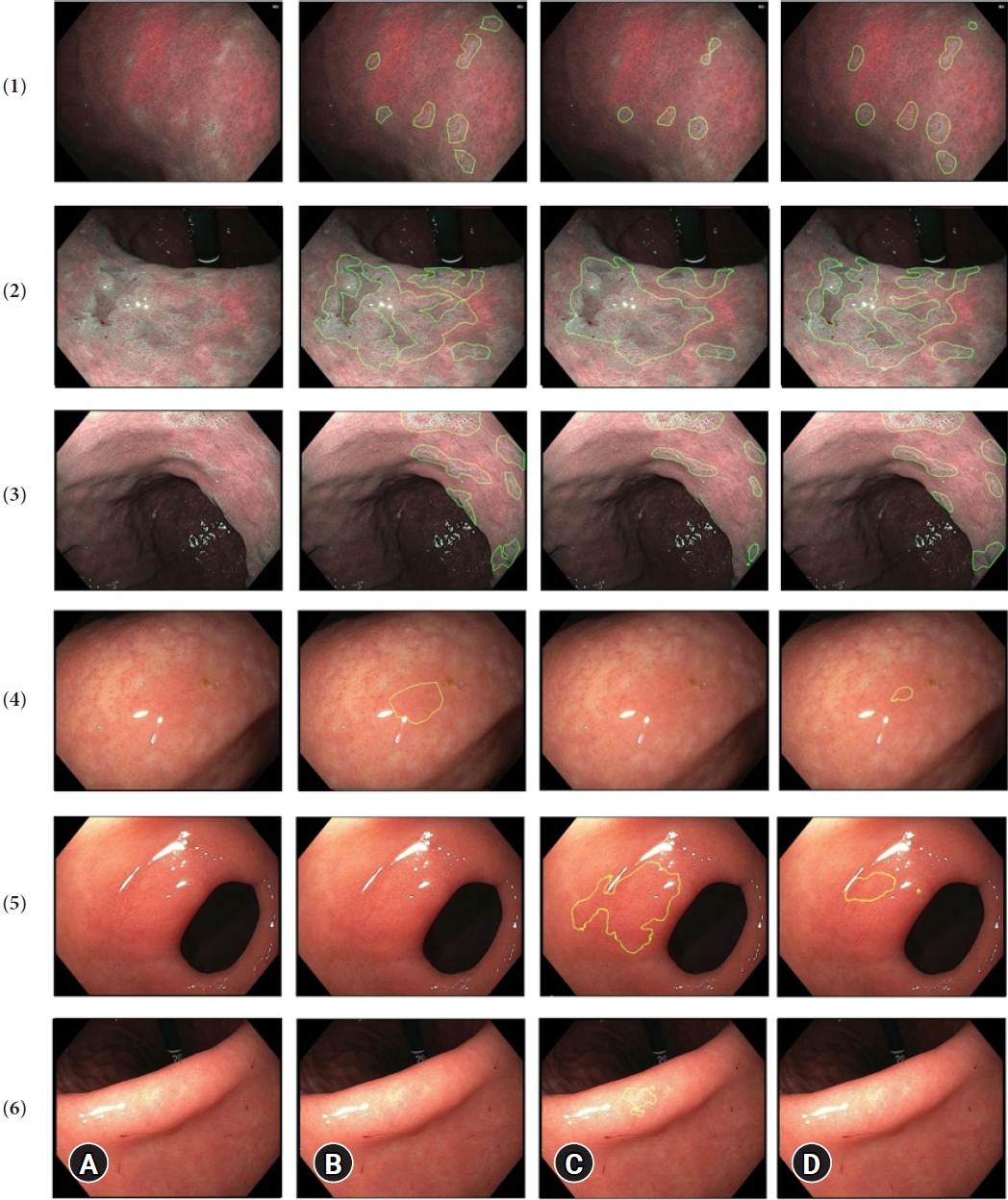
Table┬Ā1.Data separation in the gastric intestinal metaplasia dataset
Table┬Ā2.GIM detection performance of two baseline models (DeepLabV3+ and U-Net) compared to four BiSeNet variations Values are presented as percentage. GIM, gastric intestinal metaplasia; WLE, white light endoscopy; NBI, narrow-band imaging; BiSeNet, bilateral segmentation network; TL, transfer learning; CLAHE, contrast-limited adaptive histogram equalization; AUG, augmentation; PPV, positive predictive value; NPV, negative predictive value. Table┬Ā3.GIM detection performance of two baseline models (DeepLabV3+ and U-Net) compared to four BiSeNet variations, all using WLE images Values are presented as percentage. GIM, gastric intestinal metaplasia; WLE, white light endoscopy; BiSeNet, bilateral segmentation network; TL, transfer learning; CLAHE, contrast-limited adaptive histogram equalization; AUG, augmentation; PPV, positive predictive value; NPV, negative predictive value. Table┬Ā4.GIM detection performance of two baseline models (DeepLabV3+ and U-Net) compared to four BiSeNet variations, all using NBI images Values are presented as ┬▒95% confidence interval. GIM, gastric intestinal metaplasia; NBI, narrow-band imaging; BiSeNet, bilateral segmentation network; TL, transfer learning; CLAHE, contrast-limited adaptive histogram equalization; AUG, augmentation; PPV, positive predictive value; NPV, negative predictive value. Table┬Ā5.The segmentation performance of two baselines (DeepLabV3+ and U-Net) compared to four BiSeNet family models Values are presented as ┬▒95% CI. BiSeNet, bilateral segmentation network; WLE, white light endoscopy; NBI, narrow-band imaging; mIoU, mean intersection over union; GIM, gastric intestinal metaplasia; CI, confidence interval; TL, transfer learning; CLAHE, contrast-limited adaptive histogram equalization; AUG, augmentation. Table┬Ā6.The inference speed of the two benchmarks (DeepLabV3+ and U-Net) compared to four BiSeNet family model variations REFERENCES2. Lim JH, Kim N, Lee HS, et al. Correlation between endoscopic and histological diagnoses of gastric intestinal metaplasia. Gut Liver 2013;7:41ŌĆō50.
3. Panteris V, Nikolopoulou S, Lountou A, et al. Diagnostic capabilities of high-definition white light endoscopy for the diagnosis of gastric intestinal metaplasia and correlation with histologic and clinical data. Eur J Gastroenterol Hepatol 2014;26:594ŌĆō601.
4. Dixon MF, Genta RM, Yardley JH, et al. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol 1996;20:1161ŌĆō1181.
5. Ang TL, Pittayanon R, Lau JY, et al. A multicenter randomized comparison between high-definition white light endoscopy and narrow band imaging for detection of gastric lesions. Eur J Gastroenterol Hepatol 2015;27:1473ŌĆō1478.
6. Wu C, Namasivayam V, Li JW, et al. A prospective randomized tandem gastroscopy pilot study of linked color imaging versus white light imaging for detection of upper gastrointestinal lesions. J Gastroenterol Hepatol 2021;36:2562ŌĆō2567.
7. Savarino E, Corbo M, Dulbecco P, et al. Narrow-band imaging with magnifying endoscopy is accurate for detecting gastric intestinal metaplasia. World J Gastroenterol 2013;19:2668ŌĆō2675.
8. Pittayanon R, Rerknimitr R, Wisedopas N, et al. The learning curve of gastric intestinal metaplasia interpretation on the images obtained by probe-based confocal laser endomicroscopy. Diagn Ther Endosc 2012;2012:278045.
9. Sun M, Zhang G, Dang H, et al. Accurate gastric cancer segmentation in digital pathology images using deformable convolution and multi-scale embedding networks. IEEE Access 2019;7:75530ŌĆō75541.
10. Li Y, Xie X, Liu S, et al. GT-Net: a deep learning network for gastric tumor diagnosis. In: 2018 IEEE 30th International Conference on Tools with Artificial Intelligence (ICTAI); 2018 Nov 5ŌĆō7; Volos, Greece. p. 20ŌĆō24.
11. Chen LC, Zhu Y, Papandreou G, et al. Encoder-decoder with atrous separable convolution for semantic image segmentation. In: Computer Vision (ECCV 2018); p. 833ŌĆō851.
12. Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention (MICCAI 2015); p. 234ŌĆō241.
13. Read P, Meyer MP. Restoration of motion picture film. Oxford: Butterworth-Heinemann; 2000.
14. Rodriguez-Diaz E, Baffy G, Lo WK, et al. Real-time artificial intelligence-based histologic classification of colorectal polyps with augmented visualization. Gastrointest Endosc 2021;93:662ŌĆō670.
15. Wang C, Li Y, Yao J, et al. Localizing and identifying intestinal metaplasia based on deep learning in oesophagoscope. In: 2019 8th International Symposium on Next Generation Electronics (ISNE); 2019 Oct 9ŌĆō10; Zhengzhou, China. p. 1ŌĆō4.
16. Sun X, Zhang P, Wang D, et al. Colorectal polyp segmentation by U-Net with dilation convolution. In: 2019 18th IEEE International Conference on Machine Learning and Applications (ICMLA); 2019 Dec 16ŌĆō19; Boca Raton, FL. p. 851ŌĆō858.
17. Yosinski J, Clune J, Bengio Y, et al. How transferable are features in deep neural networks? In: NIPS 14: Proceedings of the 27th International Conference on Neural Information Processing Systems; 2014 Dec 8ŌĆō13; Montreal, Canada. p. 3320ŌĆō3328.
18. Russell BC, Torralba A, Murphy KP, et al. LabelMe: a database and web-based tool for image annotation. Int J Comput Vis 2008;77:157ŌĆō173.
19. Bernal J, Sanchez FJ, Fern├Īndez-Esparrach G, et al. WM-DOVA maps for accurate polyp highlighting in colonoscopy: validation vs. saliency maps from physicians. Comput Med Imaging Graph 2015;43:99ŌĆō111.
20. Jha D, Smedsrud PH, Riegler MA, et al. Kvasir-SEG: a segmented polyp dataset. In: MultiMedia Modeling: 26th International Conference, MMM 2020; 2020 Jan 5ŌĆō8; Daejeon, Korea. p. 451ŌĆō462.
21. Zuiderveld K. Contrast limited adaptive histogram equalization. In: Heckbert PS, editor. Graphics gems IV. San Diego (CA): Academic Press; 1994. p. 474ŌĆō485.
22. Yu C, Wang J, Peng C, et al. BiSeNet: bilateral segmentation network for real-time semantic segmentation. In: Computer Vision-ECCV 2018; p. 334ŌĆō349.
23. Lecun Y, Bottou L, Bengio Y, et al. Gradient-based learning applied to document recognition. Proc IEEE 1998;86:2278ŌĆō2324.
24. Goncalves W, Dos Santos M, Lobato F, et al. Deep learning in gastric tissue diseases: a systematic review. BMJ Open Gastroenterol 2020;7:e000371.
25. Mori Y, Neumann H, Misawa M, et al. Artificial intelligence in colonoscopy: now on the market. WhatŌĆÖs next? J Gastroenterol Hepatol 2021;36:7ŌĆō11.
26. Hirasawa T, Aoyama K, Tanimoto T, et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer 2018;21:653ŌĆō660.
27. Li L, Chen Y, Shen Z, et al. Convolutional neural network for the diagnosis of early gastric cancer based on magnifying narrow band imaging. Gastric Cancer 2020;23:126ŌĆō132.
28. Zhang L, Zhang Y, Wang L, et al. Diagnosis of gastric lesions through a deep convolutional neural network. Dig Endosc 2021;33:788ŌĆō796.
29. Suzuki H, Yoshitaka T, Yoshio T, et al. Artificial intelligence for cancer detection of the upper gastrointestinal tract. Dig Endosc 2021;33:254ŌĆō262.
30. Xu M, Zhou W, Wu L, et al. Artificial intelligence in the diagnosis of gastric precancerous conditions by image-enhanced endoscopy: a multicenter, diagnostic study (with video). Gastrointest Endosc 2021;94:540ŌĆō548.
31. He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27ŌĆō30; Las Vegas, NV. p. 770ŌĆō778.
32. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. In: 2015 International Conference on Learning Representations (ICLR); 2015 May 7ŌĆō9; San Diego, CA.
33. Huang G, Liu Z, van der Maaten L, et al. Densely connected convolutional networks. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017 Jul 21ŌĆō26; Honolulu, HI. p. 2261ŌĆō2269.
34. Tan M, Le QV. EfficientNet: rethinking model scaling for convolutional neural networks. In: 36th International Conference on Machine Learning (ICML 2019); 2019 Jun 9ŌĆō15; Long Beach, CA. p. 6105ŌĆō6114.
35. ASGE Technology Committee, Abu Dayyeh BK, Thosani N, et al. ASGE Technology Committee systematic review and meta-analysis assessing the ASGE PIVI thresholds for adopting real-time endoscopic assessment of the histology of diminutive colorectal polyps. Gastrointest Endosc 2015;81:502; e1ŌĆō502.e16.
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
