AbstractBarrettŌĆÖs esophagus is associated with an increased risk of adenocarcinoma. Thorough screening during endoscopic surveillance is crucial to improve patient prognosis. Detecting and characterizing dysplastic or neoplastic BarrettŌĆÖs esophagus during routine endoscopy are challenging, even for expert endoscopists. Artificial intelligence-based clinical decision support systems have been developed to provide additional assistance to physicians performing diagnostic and therapeutic gastrointestinal endoscopy. In this article, we review the current role of artificial intelligence in the management of BarrettŌĆÖs esophagus and elaborate on potential artificial intelligence in the future.
INTRODUCTIONGastroesophageal reflux disease, family history, age, or sex predisposes to BarrettŌĆÖs esophagus (BE), a complication in which stratified esophageal squamous epithelium at the level of the gastroesophageal junction is replaced by metaplastic columnar epithelium.1,2 BE is a precursor to esophageal adenocarcinoma (EAC).3 The incidence of EAC has increased significantly in recent years4,5 and is often associated with poor prognosis due to delayed diagnosis.3 Risk factors for the progression of BE to dysplastic BE or EAC include the length of the BE segment, age, ethnicity, lifestyle, and medication.6,7
The gold standard for diagnosing BE and BarrettŌĆÖs esophagus-related neoplasia (BERN) is endoscopic evaluation with histological confirmation. However, differentiating between non-dysplastic BE and BERN can be challenging, even for expert endoscopists. Existing biopsy strategies are suboptimal, with EAC and BE miss rates of >20% and 50%, respectively.8,9 This is partly attributable to poor compliance with existing biopsy protocols and the complexity of differentiation between non-dysplastic BE and BERN during endoscopic evaluation.8,9 Using imaging techniques such as narrow band imaging (NBI) with standardized classification systems for BE and BERN can help improve the diagnostic performance of endoscopists.10,11 Additionally, advanced imaging techniques such as chromoendoscopy with indigo carmine or acetic acid are valuable options and are recommended for high-quality assessment of BE (Fig. 1).11-14 However, implementation of advanced imaging techniques in daily practice requires extensive experience. Recently, several research groups have developed deep learning algorithms to improve the detection and characterization of BERN.
CURRENT STATUSArtificial intelligence: a brief introductionTime constraints and cost issues have led to the search for more efficient modalities for diagnosis and treatment of patients. To this end, artificial intelligence (AI) has become increasingly relevant to the field of medicine, especially for the early diagnosis of neoplasia. AI is an umbrella term for a wide range of topics, and the general idea is to solve problems using algorithms that require characteristics similar to human intelligence, such as the ability to learn. Machine learning (ML), as a subdiscipline of AI, describes algorithms employed to learn from pre-existing data. The field of AI most relevant to medicine, particularly endoscopy, is deep learning. Deep learning is a subtype of ML and describes a method that aims to solve defined problems with little to no supervision using vast amounts of data. Similar to the human brain, the applied algorithms or convolutional neural networks (CNNs) consist of numerous layers of neurons. CNNs learn to recognize certain patterns within the provided input data and produce a prediction or output.15 For example, in gastrointestinal (GI) endoscopy, the output could predict the dignity of an observed lesion and differentiate between neoplasia and non-neoplasia. This task is called computer-aided diagnosis (CADx). Meanwhile, identifying the lesion of interest is called computer-aided detection (CADe).16 One way to quantify the accuracy of an AI system during CADe is to determine the intersection over union (IOU). It compares the ground truth represented by a box with the output of the AI algorithm represented by a bounding box. IOU is the result of the division of the ŌĆ£area of overlapŌĆØ by the ŌĆ£area of unionŌĆØ.17 Another way to quantify the accuracy of object detection is the Dice coefficient (or S├ĖrensenŌĆōDice coefficient), which is calculated by dividing the area of union by the total number of pixels in the individual areas.18
Relevance of AI for diagnostic purposes in BarrettŌĆÖs esophagusDetecting high-grade dysplasia (HGD) and EAC during endoscopy is difficult and challenging. Considering the consequences of false negative results or missed lesions a ŌĆ£second opinionŌĆØ during endoscopic examination is an appealing thought.
van der Sommen et al.19 developed a pattern recognition-based AI system that could detect EAC on images of BE with a sensitivity and specificity of >80%. de Groof et al.20 developed an AI system based on a CNN that outperformed general endoscopists during an image-based trial with a sensitivity of 93% and specificity of 72%, compared to endoscopists with a sensitivity of 88% and specificity of 73%. In a follow-up study, the same group achieved a sensitivity, specificity, and accuracy of 91%, 89%, and 90%, respectively, in the differentiation between BE and BERN on high-definition white light endoscopy (HD-WLE) images.21 Similarly, several other groups have successfully differentiated BE from BERN during image-based studies. Hashimoto et al.22 managed to classify BE correctly with a sensitivity, specificity, and accuracy of 96.4%, 94.2%, and 95.4%, respectively. Furthermore, they managed to detect with an IOU of 0.3 and a mean average precision of 0.75. Iwagami et al.23 focused on an Asian population and developed an AI system that detects cancer at the esophagogastric junction with a sensitivity, specificity, and accuracy of 94%, 42%, and 66%, respectively. This study was conducted using 232 HD-WLE still images of 36 cancer and 43 non-cancer cases. The performance was compared to that of experts with a sensitivity, specificity, and accuracy of 88%, 43%, and 63%, respectively.
Ghatwary et al.24 compared the different methods used for the CNN-based development of CADe. On 100 HD-WLE still images, the single-shot multibox detector performed better (sensitivity, specificity, and F-score: 96%, 92%, and 0.94, respectively) than other region-based CNNs.
Struyvenberg et al.25 developed an AI system that could classify BE and BERN using NBI with a sensitivity, specificity, and accuracy of 88%, 78%, and 84%, respectively. Hussein et al.26 trained their CNN with BE and BERN images in HD-WLE and optical chromoendoscopy (i-scan from Pentax Hoya, Tokyo, Japan). They achieved a sensitivity, specificity, and area under the receiver operator curve of 91%, 79%, and 93%, respectively, during a classification task. In the same study, their CNN had an average Dice score of 50% during the segmentation task.26
Ebigbo et al.27 used HD-WLE images, NBI images, and texture and color enhancement imaging for training a CNN. Hence, they can offer multimodal CADe and CADx with promising results. Initial studies based on still images with HD-WLEŌĆöimages from the data of the Medical Image Computing and Computer Assisted Interventions Society demonstrated a sensitivity and specificity of 92% and 100%, respectively. In a study with an independent data set from the University Hospital of Augsburg, these results could be reproduced using HD-WLE images (sensitivity/specificity of 97%/88%) and NBI images (sensitivity/specificity of 94%/80%). Subsequently, the algorithm demonstrated its potential for real-life applications. The AI system captured images randomly off an endoscopic live stream and differentiated BE from EAC with an accuracy of 89.9%.28
During the early phase of this research field, even though several AI systems were able to demonstrate promising results in preclinical and pilot-phase clinical studies, AI algorithms were mostly only able to offer a per-image evaluation of BE and BERN. Ebigbo et al.29 were one of the first research groups to differentiate BE from BERN in 3 different imaging modalities in real time (Fig. 2).
Differentiating between T1a and T1b lesions during endoscopic examination is extremely difficult. Ebigbo et al.30 developed an algorithm that was able to differentiate T1a from T1b adenocarcinoma in an image-based pilot study on 230 HD-WLE images with a sensitivity, specificity, F1-score, and accuracy of 77%, 64%, 74%, and 71%, respectively. This performance was comparable to that of expert endoscopists. A meta-analysis that included 6 studies with 561 endoscopic images of patients with BE was published by Lui et al.31 Three studies used CNN as the backbone, whereas 3 used a non-CNN backbone. Overall, pooled sensitivity and specificity were approximately 86% to 88% and 86% to 90% respectively, demonstrating the promising potential of AI systems for detecting neoplastic lesions in BE.31 Nevertheless, although a meta-analysis generally incorporates data from different trials, comparing AI studies in the preclinical phase is particularly difficult because of the heterogeneity of data samples and the algorithms used for the various trials. This meta-analysis was published in 2020, and since then, more data have emerged on this topic.
Volumetric laser endomicroscopy (VLE) is an advanced imaging method that applies the principles of optical coherence tomography.32 In an observational study conducted by Smith et al.,33 VLE-guided biopsy improved, compared to random biopsies, the detection of BERN by 700% in cases where with other imaging methods no visual cues for neoplasia had been detected. However, the application of VLE and the interpretation of acquired information require practice and experience. Therefore, several research groups have attempted to address this problem using AI. Trindade et al.34 were one of the first to develop an AI system that could detect and demarcate previously determined characteristics of dysplasia on VLE images. In a case report of a patient with long-segment BE with no visual cues on HD-WLE or NBI and negative random biopsies, VLE-guided histological acquisition demonstrated focal low-grade dysplasia. Struyvenberg et al.35 developed an AI system with a sensitivity of 91% and specificity of 82% compared to VLE experts with a pooled sensitivity and specificity of 70% and 81%, respectively.
During a pilot clinical study with a manufactured spectral endoscope, Waterhouse et al.36 tested an AI system that could differentiate the spectra of BE from those of BERN with a sensitivity of 83.7% and a specificity of 85.5%.
Beyond preclinical and pilot phase clinical trials, several AI systems have already been approved for clinical use and are now commercially available. WISE VISION (NEC Corp, Tokyo, Japan) was developed and can differentiate between BE and BERN, and offers a visual representation of the area that has been classified as HGD or neoplastic. AI system was developed (CADU, Odin Medical Ltd, London, UK) that can differentiate BE from BERN and offers a visual representation when the observed image is deemed dysplastic.
As more researchers are currently in the preclinical phase or on the verge of clinical trials, a standardized way to ensure minimum requirements during the development process and later in terms of performance is urgently needed. The American Society for Gastrointestinal Endoscopy developed a guideline, the Preservation and Incorporation of Valuable Endoscopic Innovations, for the integration of new imaging technology in the context of BE. For technologies that intend to replace random biopsies with targeted biopsies, a minimum performance of 90% sensitivity, 98% negative predictive value, and 80% specificity for HGD or EAC is recommended.37 In addition to standardized threshold performance requirements, guidelines that ensure quality standards during the developmental process of AI systems are urgently needed (Table 1).19-23,25-28,30,35,36
Computer-aided quality control of upper gastrointestinal endoscopyAI has the potential to improve various aspects of GI endoscopy such as inter-examiner variability. For example, Pan et al.38 developed an AI system that automatically identifies the squamousŌĆōcolumnar junction and gastroesophageal junction on images. Ali et al.39 worked on an AI system that automatically determined the extension of the BE according to the Prague classification. With the extension of BE as a relevant factor for risk stratification, AI-assisted standardized and automated reporting has the potential to significantly improve patient care.
Additionally, a complete examination with thorough inspection is crucial to avoid missed lesions. The European Society of Gastrointestinal Endoscopy and British Society of Gastroenterology recommend photo documentation of specific landmarks during upper GI endoscopy.40,41 According to the European Society of Gastrointestinal Endoscopy, a photo documentation rate of Ōēź90% is recommended to meet the minimum quality requirements for upper GI endoscopy.41 Incomplete examinations during upper GI endoscopy can lead to an increased cancer miss rate. AI applications have the potential to provide immediate feedback on the quality of endoscopic examinations. Wu et al.42 developed an AI system, WISENSE, that can detect blind spots, document examination time, and automatically record images for photo documentation during the procedure. A randomized controlled trial compared upper GI endoscopy with or without the support of WISENSE and demonstrated a lower blind spot rate in the group that received support from the AI system (Table 2).38,39,42
LimitationsDespite currently several research teams being on the verge of clinical studies and real-life applications, their good results from preclinical studies are often limited to data from their own centers. This is particularly relevant when AI systems are commercially available and used at different centers. Moreover, during development, a heterogeneous set of data with not only a tremendous number of frames but also a sufficient number of different cases is important for the robustness of an AI system.
Furthermore, to date, no standardized method for evaluating the performance of an AI system has been created, thus indicating an urgent need to establish standardized evaluation methods. This also includes uniform terminology when describing the methods and results of the respective studies.
CONCLUSIONSModern medicine, with its ever-growing complexity coupled with limited human and material resources, is urgently needed for more efficient workflow while maintaining a high level of patient care. AI may help to solve some of these problems. Current AI applications are not being developed to replace human physicians but to support physicians during complex diagnostic and therapeutic processes. Correct interpretation of the additional information provided by AI systems is crucial for optimal performance. HumanŌĆōcomputer interaction should be a focus during the development of AI systems as the performance is. Creating AI systems that seamlessly integrate themselves into the daily routine of examiners is important. Furthermore, offering feedback on the confidence of an AI system in its current prediction is crucial. AI systems are only as good as the data input they receive, and low-quality data result in lower diagnostic performance. Although most AI systems for BE and BERN are still in the initial and preclinical phases, the immense potential of AI in routine clinical practice is evident. In the future, AI will optimize endoscopic practice and improve long-term patient outcomes.
NOTESAuthor Contributions
Conceptualization: MM, HM, AE; Data curation: MM, HM, AE; Formal analysis: MM, HM, AE; Investigation: MM, HM, AE; Methodology: MM, HM, AE; Project administration: MM, HM, AE; Resources: MM, HM, AE; Software: MM, HM, AE; Supervision: MM, HM, AE; Validation: MM, HM, AE; Visualization: MM, HM, AE; WritingŌĆōoriginal draft: MM, HM, AE; WritingŌĆōreview & editing: MM, HM, AE.
Fig.┬Ā1.Images of BarrettŌĆÖs esophagus-related neoplasia during endoscopy with an Olympus Evis X1 system (Olympus, Tokyo, Japan) in high-definition white light endoscopy (A), narrow band imaging (B), acetic acid chromoendoscopy (C), and chromoendoscopy with indigo carmine (D). 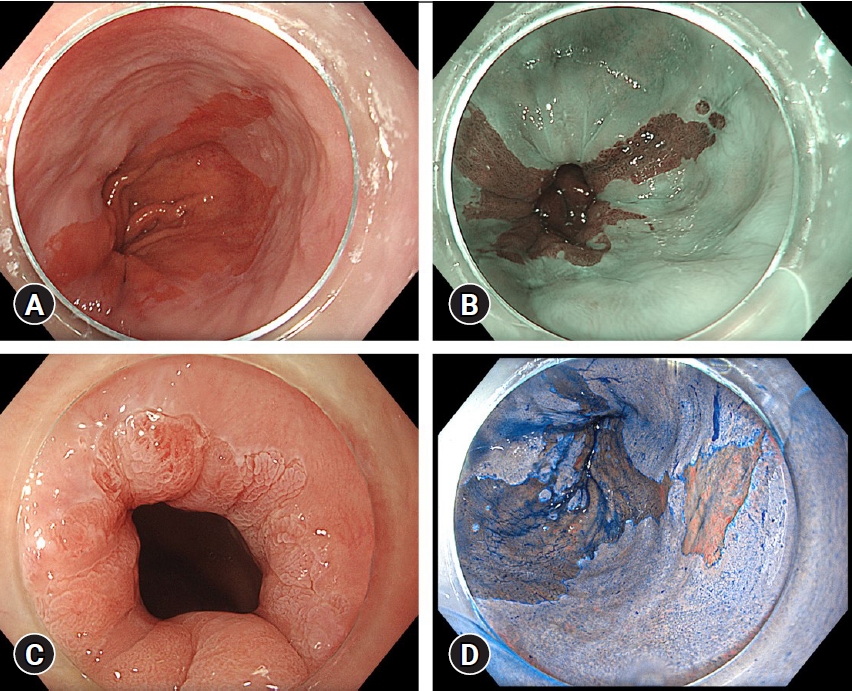
Fig.┬Ā2.Detection and characterization of BarrettŌĆÖs esophagus-related neoplasia during endoscopy with Olympus Evis X1 system using an AI system developed by the University Hospital of Augsburg and Ostbayerische Technische Hochschule Regensburg (OTH-Regensburg) with classification and segmentation in narrow band imaging (A), texture and color enhancement imaging (B) and high-definition white light endoscopy (C). The corresponding heatmaps are available at the top left corner of the user interface. 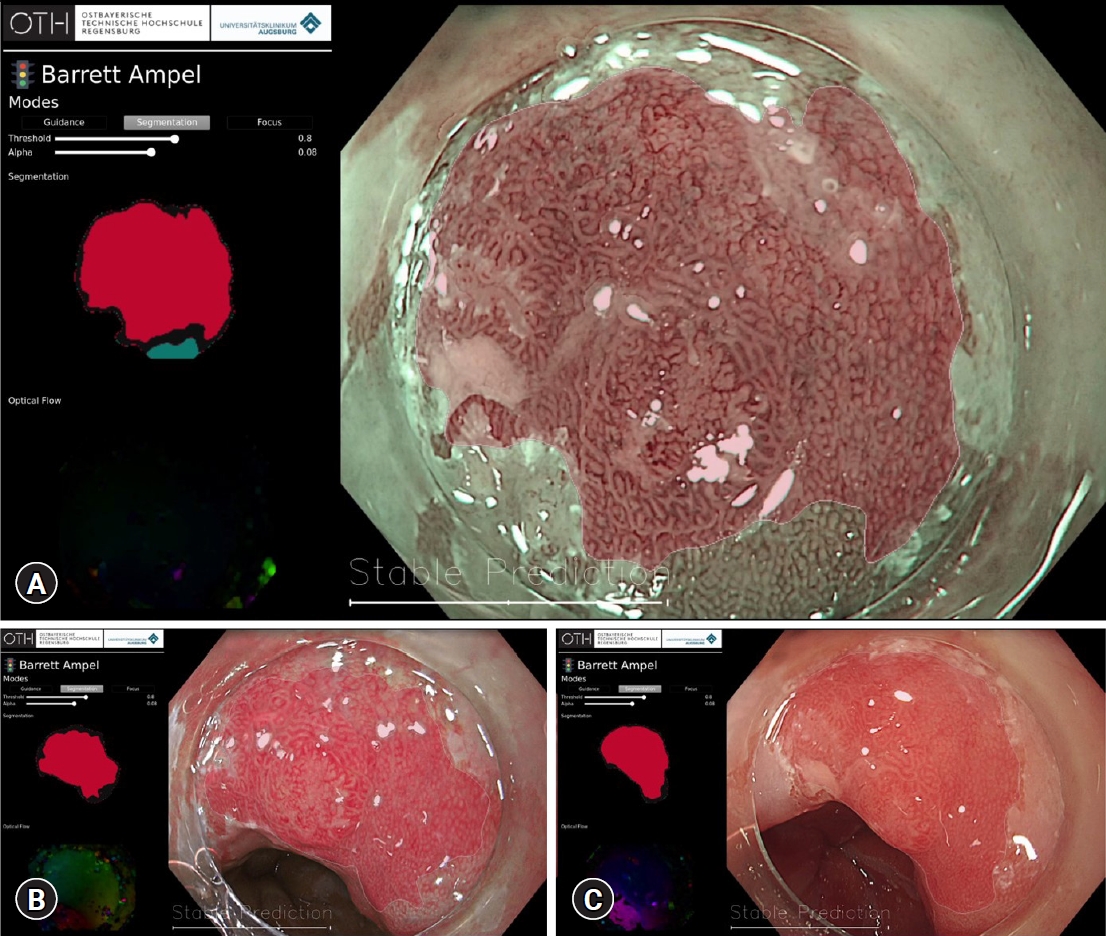
Table┬Ā1.Summary of studies exploring the application of AI during the evaluation of BarrettŌĆÖs esophagus
AI, artificial intelligence; BE, non-dysplastic BarrettŌĆÖs esophagus; HD-WLE, high-definition white light endoscopy; BERN, BarrettŌĆÖs esophagus-related neoplasia; CADx, computer-aided diagnosis; CADe, computer-aided detection; IOU, intersection over union; EGJ, esophagogastric junction; NBI, narrow band imaging; MICCAI, Medical Image Computing and Computer Assisted Interventions Society; VLE, volumetric laser endomicroscopy. Table┬Ā2.Overview of current studies in the context of computer-aided quality control during upper gastrointestinal endoscopy
REFERENCES2. Qumseya BJ, Bukannan A, Gendy S, et al. Systematic review and meta-analysis of prevalence and risk factors for BarrettŌĆÖs esophagus. Gastrointest Endosc 2019;90:707ŌĆō717.
3. Smyth EC, Lagergren J, Fitzgerald RC, et al. Oesophageal cancer. Nat Rev Dis Primers 2017;3:17048.
4. Coleman HG, Xie SH, Lagergren J. The epidemiology of esophageal adenocarcinoma. Gastroenterology 2018;154:390ŌĆō405.
5. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209ŌĆō249.
6. Kambhampati S, Tieu AH, Luber B, et al. Risk factors for progression of BarrettŌĆÖs esophagus to high grade dysplasia and esophageal adenocarcinoma. Sci Rep 2020;10:4899.
7. Chandrasekar VT, Hamade N, Desai M, et al. Significantly lower annual rates of neoplastic progression in short- compared to long-segment non-dysplastic BarrettŌĆÖs esophagus: a systematic review and meta-analysis. Endoscopy 2019;51:665ŌĆō672.
8. Visrodia K, Singh S, Krishnamoorthi R, et al. Magnitude of missed esophageal adenocarcinoma after BarrettŌĆÖs esophagus diagnosis: a systematic review and meta-analysis. Gastroenterology 2016;150:599ŌĆō607.
9. Singer ME, Odze RD. High rate of missed Barrett's esophagus when screening with forceps biopsies. Esophagus 2023;20:143ŌĆō149.
10. Sharma P, Bergman JJ, Goda K, et al. Development and validation of a classification system to identify high-grade dysplasia and esophageal adenocarcinoma in BarrettŌĆÖs esophagus using narrow-band imaging. Gastroenterology 2016;150:591ŌĆō598.
11. ASGE Technology Committee, Thosani N, Abu Dayyeh BK, et al. ASGE Technology Committee systematic review and meta-analysis assessing the ASGE Preservation and Incorporation of Valuable Endoscopic Innovations thresholds for adopting real-time imaging-assisted endoscopic targeted biopsy during endoscopic surveillance of BarrettŌĆÖs esophagus. Gastrointest Endosc 2016;83:684ŌĆō698.
12. Qumseya BJ, Wang H, Badie N, et al. Advanced imaging technologies increase detection of dysplasia and neoplasia in patients with Barrett's esophagus: a meta-analysis and systematic review. Clin Gastroenterol Hepatol 2013;11:1562ŌĆō1570.
13. Tholoor S, Bhattacharyya R, Tsagkournis O, et al. Acetic acid chromoendoscopy in BarrettŌĆÖs esophagus surveillance is superior to the standardized random biopsy protocol: results from a large cohort study (with video). Gastrointest Endosc 2014;80:417ŌĆō424.
14. Chedgy FJ, Subramaniam S, Kandiah K, et al. Acetic acid chromoendoscopy: improving neoplasia detection in BarrettŌĆÖs esophagus. World J Gastroenterol 2016;22:5753ŌĆō5760.
15. Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med 2019;25:44ŌĆō56.
16. van der Sommen F, de Groof J, Struyvenberg M, et al. Machine learning in GI endoscopy: practical guidance in how to interpret a novel field. Gut 2020;69:2035ŌĆō2045.
17. Hsiao CH, Lin PC, Chung LA, et al. A deep learning-based precision and automatic kidney segmentation system using efficient feature pyramid networks in computed tomography images. Comput Methods Programs Biomed 2022;221:106854.
18. Zou KH, Warfield SK, Bharatha A, et al. Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol 2004;11:178ŌĆō189.
19. van der Sommen F, Zinger S, Curvers WL, et al. Computer-aided detection of early neoplastic lesions in BarrettŌĆÖs esophagus. Endoscopy 2016;48:617ŌĆō624.
20. de Groof AJ, Struyvenberg MR, van der Putten J, et al. Deep-learning system detects neoplasia in patients with BarrettŌĆÖs esophagus with higher accuracy than endoscopists in a multistep training and validation study with benchmarking. Gastroenterology 2020;158:915ŌĆō929.
21. de Groof AJ, Struyvenberg MR, Fockens KN, et al. Deep learning algorithm detection of BarrettŌĆÖs neoplasia with high accuracy during live endoscopic procedures: a pilot study (with video). Gastrointest Endosc 2020;91:1242ŌĆō1250.
22. Hashimoto R, Requa J, Dao T, et al. Artificial intelligence using convolutional neural networks for real-time detection of early esophageal neoplasia in BarrettŌĆÖs esophagus (with video). Gastrointest Endosc 2020;91:1264ŌĆō1271.
23. Iwagami H, Ishihara R, Aoyama K, et al. Artificial intelligence for the detection of esophageal and esophagogastric junctional adenocarcinoma. J Gastroenterol Hepatol 2021;36:131ŌĆō136.
24. Ghatwary N, Zolgharni M, Ye X. Early esophageal adenocarcinoma detection using deep learning methods. Int J Comput Assist Radiol Surg 2019;14:611ŌĆō621.
25. Struyvenberg MR, de Groof AJ, van der Putten J, et al. A computer-assisted algorithm for narrow-band imaging-based tissue characterization in BarrettŌĆÖs esophagus. Gastrointest Endosc 2021;93:89ŌĆō98.
26. Hussein M, Gonz├Īlez-Bueno Puyal J, Lines D, et al. A new artificial intelligence system successfully detects and localises early neoplasia in BarrettŌĆÖs esophagus by using convolutional neural networks. United European Gastroenterol J 2022;10:528ŌĆō537.
27. Ebigbo A, Mendel R, Probst A, et al. Computer-aided diagnosis using deep learning in the evaluation of early oesophageal adenocarcinoma. Gut 2019;68:1143ŌĆō1145.
28. Ebigbo A, Mendel R, Probst A, et al. Real-time use of artificial intelligence in the evaluation of cancer in BarrettŌĆÖs oesophagus. Gut 2020;69:615ŌĆō616.
29. Ebigbo A, Mendel R, Probst A, et al. Multimodal imaging for detection and segmentation of BarrettŌĆÖs esophagus-related neoplasia using artificial intelligence. Endoscopy 2022;54:E587.
30. Ebigbo A, Mendel R, R├╝ckert T, et al. Endoscopic prediction of submucosal invasion in BarrettŌĆÖs cancer with the use of artificial intelligence: a pilot study. Endoscopy 2021;53:878ŌĆō883.
31. Lui TK, Tsui VW, Leung WK. Accuracy of artificial intelligence-assisted detection of upper GI lesions: a systematic review and meta-analysis. Gastrointest Endosc 2020;92:821ŌĆō830.
32. Elsbernd BL, Dunbar KB. Volumetric laser endomicroscopy in BarrettŌĆÖs esophagus. Tech Innov Gastrointest Endosc 2021;23:P69ŌĆōP76.
33. Smith MS, Cash B, Konda V, et al. Volumetric laser endomicroscopy and its application to BarrettŌĆÖs esophagus: results from a 1,000 patient registry. Dis Esophagus 2019;32:doz029.
34. Trindade AJ, McKinley MJ, Fan C, et al. Endoscopic surveillance of BarrettŌĆÖs esophagus using volumetric laser endomicroscopy with artificial intelligence image enhancement. Gastroenterology 2019;157:303ŌĆō305.
35. Struyvenberg MR, de Groof AJ, Fonoll├Ā R, et al. Prospective development and validation of a volumetric laser endomicroscopy computer algorithm for detection of BarrettŌĆÖs neoplasia. Gastrointest Endosc 2021;93:871ŌĆō879.
36. Waterhouse DJ, Januszewicz W, Ali S, et al. Spectral endoscopy enhances contrast for neoplasia in surveillance of BarrettŌĆÖs esophagus. Cancer Res 2021;81:3415ŌĆō3425.
37. Sharma P, Savides TJ, Canto MI, et al. The American Society for Gastrointestinal Endoscopy PIVI (Preservation and Incorporation of Valuable Endoscopic Innovations) on imaging in BarrettŌĆÖs Esophagus. Gastrointest Endosc 2012;76:252ŌĆō254.
38. Pan W, Li X, Wang W, et al. Identification of BarrettŌĆÖs esophagus in endoscopic images using deep learning. BMC Gastroenterol 2021;21:479.
39. Ali S, Bailey A, Ash S, et al. A pilot study on automatic three-dimensional quantification of BarrettŌĆÖs esophagus for risk stratification and therapy monitoring. Gastroenterology 2021;161:865ŌĆō878.
40. Beg S, Ragunath K, Wyman A, et al. Quality standards in upper gastrointestinal endoscopy: a position statement of the British Society of Gastroenterology (BSG) and Association of Upper Gastrointestinal Surgeons of Great Britain and Ireland (AUGIS). Gut 2017;66:1886ŌĆō1899.
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
