AbstractOver the past decade, technological advances in deep learning have led to the introduction of artificial intelligence (AI) in medical imaging. The most commonly used structure in image recognition is the convolutional neural network, which mimics the action of the human visual cortex. The applications of AI in gastrointestinal endoscopy are diverse. Computer-aided diagnosis has achieved remarkable outcomes with recent improvements in machine-learning techniques and advances in computer performance. Despite some hurdles, the implementation of AI-assisted clinical practice is expected to aid endoscopists in real-time decision-making. In this summary, we reviewed state-of-the-art AI in the field of gastrointestinal endoscopy and offered a practical guide for building a learning image dataset for algorithm development.
INTRODUCTIONArtificial intelligence (AI) is a concept that was first introduced in the 1950s and which has, in the past decade, made great strides driven by the accumulation of a large amount of data that can train ever more sophisticated AI, as well as by improved computational power leveraged through hardware innovations, including graphic processing units, and advances in deep learning (DL) technology. Various AI applications have been reported in the field of gastrointestinal (GI) endoscopy, especially with the use of DL technology, including convolutional neural networks (CNNs). Rapid advances in AI technology in recent years have increased the need for endoscopists to become familiar with AI and its data structure. This article introduces the basic concepts of AI, machine learning (ML), and DL with a focus on the clinical applications of AI in the field of GI endoscopy. Additionally, we provide guidance for building imaging datasets for developing DL models and discuss various challenges posed by this process.
MACHINE LEARNINGAI refers to the ability of a computer to perform tasks in a manner similar to human intelligence. The field has gradually grown, and is now subdivided into several areas. Among them, ML refers to a system that can learn from data without explicit programming.1 Samuel2 defined ML as āProgramming a computer to learn from experience that should eventually eliminate the need for much of this detailed programming effortā. ML is traditionally derived from pattern-recognition systems and possesses an algorithm that recognizes features or patterns associated with data to make specific predictions, with repeated practice leading to improved performance.
In contrast with traditional computer programs, ML models are not programmed with rules, but learn from examples.3 For a particular task, examples are provided in the form of inputs (features) and outputs (labels). Using a learning-from-observation algorithm, a computer can determine how to perform mapping from a label function to generate a model that can generalize information so that it can perform the task consistently with unfamiliar input features. When training the model, features are converted into expected labels through complex, multilayered mathematical functions. The algorithm determines how to value each parameter to ensure that the model accurately reflects reality and makes more precise predictions. ML can be roughly divided into supervised and unsupervised models. Supervised learning takes place by the provision of labeled data that provides the correct answer. In contrast, unsupervised methods are designed for automated clustering of similar data based on commonalities. Therefore, unsupervised learning can be used as a primary tool for identifying appropriate features following supervised learning. ML has limitations in that it can only learn from the data in the training set; as a result, the developed model might not be able to make accurate predictions if new examples arise that are different from those in the training set.
DEEP LEARNING AND CONVOLUTIONAL NEURAL NETWORKAmong the ML methods, artificial neural networks (ANNs) mimic the information processing of the human brain. Each neuron is a computing unit, and all neurons are connected to build a sophisticated network. Neurons exchange electrical information signals, and only when an input signal exceeds a threshold, the signal is transmitted to the next neuron. McCulloch and Pitts4 first proposed this concept in 1943. Over time, the theory has emerged that learning can selectively strengthen the synaptic activation between certain neurons. In this theory, each input can be multiplied by a weighting factor, and all the multiplied inputs are then summed. An advantage of ANN is that it can model highly nonlinear relationships between inputs and desired outputs by combining many neurons into layers.5 As one of the ML techniques, DL, which is based on ANN, emerged relatively quickly around 2010. In 2006, Hinton and Salakhutdinov6 named a multilayered neural network composed of several hidden layers a ādeep neural network (DNN)ā, and the learning method based on DNN was first named DL.
DL is an ML algorithm that extracts and transforms features by using multiple layers of nonlinear processes. āFeature extractionā is the process of selecting variables that are likely to have predictive power for an objective, and āTransformationā is the process of changing the data in a more effective way to build a model. Recently, its performance has been improved such that it is now possible to design ANNs with tens to hundreds of layers with ease.
In the field of medical image processing, CNNs are the most commonly used ANN structure for image analysis. CNNs are DNNs specialized in image recognition technology, which were first introduced by Le Cunās team.7 CNNs use the principles of image processing and recognition used by the brainās visual cortex. They can gradually learn high-level features through complex connections that mimic the action of the human visual cortex.8 Put more simply, the CNN model consists of three layers: (1) convolutional layer, (2) pooling layer, and (3) final classification (Fig. 1). During convolution, the kernel, an image filter of a certain size, scans the entire image and passes the output value to the next node. The next step, pooling, reduces the dimensions of the feature. Features that are effective for learning are selected by this process. A feature map presented through these processes enters the fully connected layers, and the final classification result of an image can be derived. Class activation mapping refers to the location information within an image that allows a CNN to predict a specific class, and is the output of a particular convolution layer. Activation maps can be used for the visualization of CNNs (Fig. 2).
In this era of big data, the amount of data to be analyzed using these computations is unimaginably large. With dramatic improvements in computing power and graphic processing units, more complex calculations are now possible, including in DL. AlexNet, the winner of the ImageNet Large-Scale Visual Recognition Challenge competition in 2012, is a DL algorithm with eight hidden layers. It has increased the recognition rate of conventional ML algorithms, which remained in the 70% over the past 10 years, to 85% in one significant leap.9 Microsoftās ResNet, which won the ImageNet Large-Scale Visual Recognition ChalĀlenge in 2015, has 152 deeper layers and has dramatically improved performance in various areas such as image recognition and facial recognition.10
CNN models are being applied to images and medical data analysis in various areas.11,12 AI in medical imaging has been investigated in several fields, including radiology, neurology, orthopedics, pathology, and gastroenterology. Since the development of AlexNet in 2012, DL in the image recognition field has mostly utilized CNNs. Although it is not yet a top player in medical imaging, image recognition studies in GI endoscopy are actively underway and have surprisingly good potential. In the next section, we introduce various cases in which DL is used in GI endoscopy.
ARTIFICIAL INTELLIGENCE IN GASTROINTESTINAL ENDOSCOPYML techniques are used in various areas of GI endoscopy. Since adenoma detection rates (ADRs) during colonoscopy and medical decision-making regarding detected polyps vary greatly depending on the experience and skill of the endoscopist, several studies have introduced computer-assisted diagnostic techniques that can reduce variability between endoscopists and potentially improve ADRs (Table 1).13-31
Since the doctor who performs the endoscopy is a human, simple errors can occur due to fatigue caused by overwork and/or special conditions. The rate of polyps missed on colonoscopy is reportedly as high as 25%.32 Certain types of polyps are more likely to be missed and may result in progression to cancer. An enormous advantage of endoscopic diagnosis using AI is that polyp detection or, theoretically, the decision-making process, is not susceptible to fatigue-related errors.
The application of the CNN algorithm to polyp detection appears to be very promising. Billah et al.13 utilized images from a public dataset to develop an algorithm that achieved a polyp detection sensitivity of 98% to 99%. The automated detection of polyps outperformed the endoscopist by 86% to 74% in terms of accuracy.14 Despite their high false-positive rates, all of these studies showed promising results on the use of CNN to identify colon polyps. For trainee endoscopists just starting to learn colonoscopy, computer-assisted diagnostic technologies employing AI can be a significant help, not only for polyp detection, but also for characterizing the detected polyp. This improves ADRs and leads to appropriate decision-making regarding follow-up examinations and treatment plans.
The ultimate goal of AI algorithms in the field of colonoscopy is real-time detection of polyps. Real-time detection should exhibit high sensitivity and specificity and provide practical information to the operator without interfering with the operation. In addition, appropriate hardware performance for the AI system should be available to ensure low latency time between polyp detection and screen display.
Urban et al.33 developed an ImageNet-based CNN algorithm and validated it using 11 colonoscopy videos. The algorithmās performance yielded a sensitivity of 97% and a specificity of 96%, executing at 10 microseconds per frame. Using narrow-band image (NBI) video frames and videos, the algorithm developed by Byrne et al.15 showed a sensitivity of 98%, a specificity of 83%, and an accuracy of 94% for 125 videos of 106 polyps. Recently, Repici et al.16 performed a randomized trial with 685 subjects undergoing colonoscopy, using a real-time polyp detection algorithm. The computer-aided detection system improved the ADR from 40.4% to 54.8%, particularly for diminutive polyps (<5 mm), but did not increase the physicianās withdrawal time.
AI technology can also be used to characterize the detected polyps. One study has shown excellent performance of AI technology in discriminating whether the detected polyp is an adenomatous polyp, which requires removal, a hyperplastic polyp, which does not require removal, and any additional pathological features.17 In cases where a resected polyp is considered to have low clinical significance and may not require additional pathological review, real-time histological AI confirmation may help clinicians avoid unnecessary pathological verification of the resected polyp. This strategy can reduce the cost of unnecessary pathological examinations.
While colon polyps are lesions with clear borders, target lesions in the upper GI tract are generally subtle and difficult to find.34 Surprisingly, the difference between well-trained AI and a general endoscopist may be even greater in this scenario. Several studies have been published on the probability-based detection of suspicious gastric tumor lesions and the detection of Helicobacter pylori-infected gastric mucosa.20,21 For detecting neoplastic lesions in the stomach, AI achieved a pooled area under the curve of 0.96, with a pooled sensitivity of 92.1% and specificity of 95.0% in a recent meta-analysis,35 superior to the efficacy of endoscopists. On the other hand, AI has some difficulties detecting and discriminating between neoplastic lesions of the esophagus, and this was particularly noticeable for Paris type 0āIIb (superficial-flat) lesions.36 Special imaging techniques such as NBI are helpful for developing AI algorithms to detect such esophageal lesions.35
Endocytoscopy (H290ECI; Olympus, Tokyo, Japan) is a newly introduced in vivo contact-type microscopic imaging modality that provides real-time cellular-level images during endoscopy.37,38 A multicenter study in Japan22 has validated the diagnostic efficacy of EndoBRAIN (Cybernet Systems Co., Tokyo, Japan), an AI-based system that analyzes cell nuclei, crypt structures, and microvessels in endoscopic images during the identification of colonic neoplasms. EndoBRAIN identified colonic lesions with 96.9% sensitivity and 100% specificity when pathology findings were used as the gold standard.
Small-bowel capsule endoscopy is another field that is expected to benefit from advances in AI pattern-recognition technology. Since capsule endoscopy interpretation is a relatively tedious, time-consuming, and error-prone process, AI algorithms are highly likely to be used in clinical practice in the near future.39 Despite the high necessity and high expectations, training a reliable AI algorithm for capsule endoscopy has faced many obstacles, foremost among which are the low resolution of capsule endoscopy images and blurred pictures that are randomly taken during passive movement through the small bowel. Image enhancement technologies are promising tools for overcoming obstacles in developing an effective capsule endoscopy AI. āImage enhancingā is designed to maximize the efficacy of AI-learning by generating optimized images for learning from existing suboptimal images. Several image-enhancing methods have been developed, including three-dimensional (3D) image reconstruction, chromo-endomicroscopy, and image resolution improvement software for denoising and de-blurring. For example, the efficacy of 3D reconstruction with dual-camera capsules (MiroCam MC 4000; IntroMedic, Seoul, Korea) was prospectively validated and appeared to be exceptionally useful in the characterization of subepithelial tumors.40 In a recent multicenter retrospective study,28 a CNN outperformed human gastroenterologists in multiple lesion detection with capsule endoscopy (sensitivity, 99.88% vs. 74.57%; p<0.001). The CNN performed the task in 5.9 minutes on average, while endoscopists required a manual reading time of 96.6 minutes. Active locomotion capsule endoscopy is used to control the capsuleās position freely using magnetic force, along with automated reading by an AI system, which is also in development.41
Automating the endoscopic assessment of ulcerative colitis (UC) is another area of AI research.42 Several recent studies have reported a CNN system to assess endoscopic severity in UC.43,44 Ozawa et al.45 evaluated a CNN-based algorithm for endoscopically classifying disease activity in 841 patients with UC. The system selectively pointed out Mayo class 2ā3 images severely affected by UC with an area unĀder the curve of 0.86 (95% confidence interval, 0.84ā0.87).
CONSIDERATIONS FOR THE BUILDING OF AN AI-LEARNING DATASETThe establishment of separate training, validation and test setsDatasets of acquired images should be divided according to their purpose,46 which are usually training, validation, and testing. The training set is used to learn specific patterns and their labels from images.47 This set is used when training an ML model by iteratively updating model parameters until the model best fits the data. The validation set is sometimes used interchangeably with a tuning set. After learning a specific pattern, it is used to verify whether the model is underfitting or overfitting. The āhyperparametersā of the model are then tuned. In medical research, models should be validated using datasets that are completely independent of training or validation sets. The test set is used to evaluate the modelās performance before applying the ML model in clinical practice. Test sets cannot be used to train or tune ML models, including hyperparameters or ML method selection. The test set is used to avoid selection bias and to report unbiased predictions once the design of the model is decided based on the performance of the validation set.47
It is also necessary to consider how to proportionally separate the entire image data into each category. The basic rule is that the validation and test sets must be sufficiently large to reflect real-world variability. The remaining data are then distributed to the training set. If the size of the total data is small, it is appropriate to distribute a relatively large ratio to the validation and testing sets compared with the training set. By contrast, if the data size is large, a greater weight can be used for the training set.
Sample size and class imbalance problemsIt is important to ensure sufficient sample size. It is desirable to include a large dataset with various features for training and validation.48 The optimal sample size may vary depending on the task. DL requires a larger sample size than traditional ML methods to optimize performance. This is necessary to minimize the risk of overfitting, which will be discussed later. Using data from multiple sources, rather than data from a single data storage, can reduce the sample bias and increase the generalizability of the algorithm. To obtain images, one can also consider using many open-source datasets.
However, in practice, determining the amount of data required should take several complicating factors into consideration, including task difficulty, input data types, and quality of labels of the work.47 In reality, the cost of obtaining and labeling data is sometimes considered as important as the design of the AI model.
Class imbalance is often a problem in real data, occurring when there are large differences in the amount of data possessed by each category. For example, if a researcher collects images of cases with lesions and data from subjects without the disease, lesions appear infrequently and in many different conditions; therefore, collecting sufficient images for certain diagnoses is not easy. Most āreal-worldā clinical data have a class imbalance problem. Techniques for class balancing in DL include āweight balancingā and āover and undersamplingā. Sampling is a simple method that a general researcher can perform without the need for a computer engineer. There is an undersampling method that selects only a portion of the majority class and a method of oversampling makes as much data as possible available by making multiple copies of the minority class.
The problem of overfittingOverfitting occurs when the ML model is overtrained on the training data itself, and is not generalizable to new datasets. In theory, if a sufficiently large number of parameters are input into a mathematical model, any dataset can fit the model. Such models may not perform well clinically when their fit depends on these additional variables. If the model is applied to a dataset that differs from the training set, prediction may fail.
Techniques such as reducing the number of parameters in a model or preventing a model from overfitting a dataset are collectively called "regularization". An example is the smoothing of a noisy curve. Regularization techniques include ensemble, data augmentation, early stopping, fine-tuning, warm start, and parameter regularization. āEnsembleā is a technique to improve the stability of the final prediction by combining multiple outputs of a ML model. This is achieved by averaging the output with the same input data.
Most regularization techniques affect the learned parameters of ML models. Prior to using these techniques, additional hyperparameters must be set. When randomness is controlled during ML training, modifying the hyperparameter settings completely determines the final values of the learned parameters. Similarly, changing the hyperparameters and training a new ML model can change the values of the learned parameters. Because hyperparameters have a large impact on model performance, tuning them is important in ML research.
Data preparation, curation, and annotationThe quality of the reference training material is the most important factor in determining the performance of a model. However, the determination of reference standards is often subjective and can lead to interobserver variability. This variability can be reduced through adjudication by an experienced panel of experts. In addition, high-quality reference standards are important for demonstrating the model performance. To avoid bias, the reference standards should be determined independently. Clinicians involved in grading images should be blinded to ML predictions. Even a small difference in the model performance can potentially affect many patients.
A diversity of images and lack of bias have been suggested as essential requirements for building a high-quality image database. The lack of these features can skew the algorithmās ability to correctly categorize input variables.49 In addition, different models trained with datasets independently generated by various institutions might present low predictive power when applied to heterogeneous groups. Such isolated data are termed ādatabase islandsā. The advent of artificial image augmentation technologies, such as generative adversarial networks that perform flipping, cropping, resizing, and blurring of existing images, have been introduced and utilized to diversify and increase the number of datasets.
Problems with incomplete data, duplicate data, and other dataset formats must be addressed. This can be time-consuming and labor-intensive, particularly for large datasets. Learning materials can be independently labeled by consensus to establish a reference standard or āground truthā. In addition, one may need to collect additional data to confirm the pathology findings and medical records. To optimize efficiency at this stage, it can be useful to identify the variables that are most relevant to the goal.
Annotating secured video or image data is labor-intensive. Several methods of annotation are employed, including bounding boxes, polygonal segmentation, semantic segmentation, 3D cuboids, lines, and splines. Sharing a standardized annotation tool and common reference standards among readers participating in this process is important to ensure the quality of the data and to save time (Fig. 3). Because a small dataset is usually not adequate to achieve sufficient AI performance, multiple organizations should share large-scale datasets and perform joint annotation. To this end, there has been ongoing discussion on whether to build a large public dataset suitable for each healthcare community. As an example of this type of effort, Ding et al.28 collected 108 capsule endoscopy cases from 77 medical centers in 2019. They labeled and annotated over 158,000 images as either normal or as one of 10 abnormal categories for training. They used more than 100 million images for validation and developed a multiple-lesion-detecting ResNet-based algorithm for capsule endoscopy reading.
Data storage and federated learningData storage and training of the model can be performed on local hardware or remotely on a cloud-computing platform. Unified learning is a new technique that can train a single algorithm using data from multiple distributed devices, such as multiple local servers, at different sites, without the need to exchange or transmit images to a central repository.
Federated learning (FL) is a new learning paradigm that allows developers and organizations to train DNNs using distributed training data from multiple locations. FL has two significant advantages: data privacy and communication efficiency. In particular, FL has gained increasing attention from healthcare AI developers. FL makes it possible to use clinical data from hospitals for learning without fear of breaching patient confidentiality, so that developers can easily overcome legal and ethical barriers. In addition, FL can reduce the network traffic burden as the optimized model is redistributed by sharing only the necessary information, without sharing all clinical data directly. Block chain technology is expected to be used in conjunction with FL to record audit trails in the future.
Ethical issuesEthical issues cannot be avoided when constructing AI datasets. When attempting to use endoscopy results as learning data, it is necessary to obtain the consent of individual patients. Therefore, wording should be inserted into the consent form prior to endoscopy, and the patient should be notified in advance. If a patient refuses to permit his or her endoscopy results to be used for AI-learning data, they cannot be integrated into the learning dataset.
When constructing an infrastructure to share datasets between multiple institutions, processes such as de-identification and encryption are required. Clinical data transmitted to other institutions should not contain data that would allow patient identification. As data become accessible to many subjects from multiple institutions, the potential for invasion of individual privacy will increase significantly. In addition, the security and encryption of data transmitted between institutions are also important. The nature of data makes it a potential resource that can be directly linked to economic value. Thus, these data can be considered vulnerable to cyber-attack.
FUTURE PERSPECTIVESThe most important aspect in developing an AI model with good predictive power is the construction of a large-scale, high-quality dataset. Unfortunately, such high-quality datasets are still scarce. There is an old adage in computer science, āgarbage in, garbage outā, which is still valid even at the cutting-edge of modern computational power. It is difficult to build a dataset suitable for AI training at a single institution. Owing to the development of information and communication technologies, an infrastructure that can share a large amount of data is easily built. However, the foundation of a high-quality dataset that can be shared and utilized by multiple organizations is an important factor in accelerating the development of AI models that can exhibit the required performance levels. To ensure this, the legal regulations surrounding data sharing should be improved and a control tower should promote cooperation between institutions. The introduction of AI technology is expected to lead to an increase in medical expenses. In this regard, social consensus among doctors, patients, and insurance companies is necessary.50
As of 2021, commercialized computer-aided diagnosis systems such as EndoBRAIN-EYE (Cybernet Systems Co.), DISCOVERY (PENTAX Medical, Tokyo, Japan), and the GI Genius module (Medtronic, Minneapolis, MN, USA) have been introduced. These technologies support endoscopic diagnosis and are certified in Europe51; however, issues remain, as we do not know whether these computer-aided diagnostic technologies reduce the long-term incidence of GI diseases, mortality rate, or overall medical expense. In addition, these computer-aided diagnosis systems use their own isolated datasets for training, and are therefore not fully validated in real-world practice. The current evidence is based on retrospective studies, and is prone to a high risk of investigator-induced bias. Future prospective multicenter studies are mandatory before US Food and Drug Administration approval and widespread use. Currently, there are only a few prospective studies in this field. In particular, in Korea, a clinical prospective study is possible only through strict legal procedures for the medical use of the developed AI algorithm. AIās multi-diagnosis accuracy and comprehensiveness have not yet reached this point, and many prospective clinical studies can be conducted only after overcoming these technical and legal issues.
Privacy is an important aspect of AI. It is well known that a large amount of data is needed to train AI. However, most of these data include personal information. Personal information should be pseudonymized and used for learning. All external features, such as the eyes, nose, and mouth in the photographed images, should be deleted before use. Complex pseudonymization is required, depending on the type of information, such as information removal. For this purpose, privacy-preserving machine-learning technology is also being actively investigated.
CONCLUSIONSIt is evident that, in the near future, the implementation of AI for GI endoscopy will benefit clinicians in various ways. From traineesā education to real-time microscopic interpretation, computerized algorithms may serve as faithful assistants in the field of endoscopy. However, a sufficient amount of high-quality data is essential for the successful development of AI. The lack of learning data may explain why it has not yet reached clinical use in GI endoscopy. Engineers, endoscopists, and physicians should therefore understand how to build learning datasets for AI training. In the near future, we anticipate that we will achieve the goal of developing strong AI algorithms through the construction of multi-institutional, high-efficiency learning materials in tandem with the latest developments in cloud computing and FL.
NOTESFig.Ā 2.Class activation map of a capsule endoscopy image. (A) Erosions with depression of the mucosa are highlighted in red. (B) Detection of vascular lesions on the small-bowel mucosa is visualized in a class activation map. 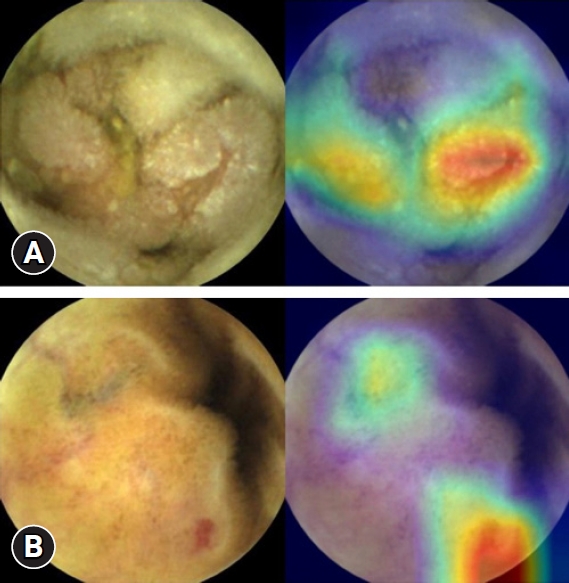
Fig.Ā 3.Annotation process of capsule endoscopy images for the development of convolutional neural networks. Three categorial numbers will be applied to each image in terms of medical significance, degree of protrusion and type of lesion (e.g., vascular, inflammatory, polypoid) according to a predefined reference standard. 
TableĀ 1.Published studies on the application of artificial intelligence in the field of gastrointestinal endoscopy
NICE, Narrow-band Imaging International Colorectal Endoscopic; NPV, negative predictive value; PPV, positive predictive value; AUROC, area under the receiver operating characteristic curve; EGD, esophagogastroduodenoscopy; EGC, early gastric cancer; M-NBI, magnifying endoscopy with narrow-band imaging; DCNN, deep convolutional neural network. REFERENCES1. Ruffle JK, Farmer AD, Aziz Q. Artificial intelligence-assisted gastroenterology- promises and pitfalls. Am J Gastroenterol 2019;114:422ā428.
2. Samuel AL. Some studies in machine learning using the game of checkers. IBM J Res Dev 1959;3:210ā229.
4. McCulloch WS, Pitts W. A logical calculus of the ideas immanent in nervous activity. 1943. Bull Math Biol 1990;52:99ā115.
5. Litjens G, Ciompi F, Wolterink JM, et al. State-of-the-art deep learning in cardiovascular image analysis. JACC Cardiovasc Imaging 2019;12:1549ā1565.
6. Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science 2006;313:504ā507.
7. Le Cun Y, Boser B, Denker JS, et al. Handwritten digit recognition with a back-propagation network. In: Touretzky DS, editor. Advances in neural information processing systems 2. San Francisco (CA): Morgan Kaufmann Publishers Inc.; 1990. p. 396ā404.
8. Ahmad OF, Soares AS, Mazomenos E, et al. Artificial intelligence and computer-aided diagnosis in colonoscopy: current evidence and future directions. Lancet Gastroenterol Hepatol 2019;4:71ā80.
9. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM 2017;60:84ā90.
10. He K, Zhang X, Ren S, et al. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 Jun 27-30; Las Vegas, NV. p. 770ā778.
11. Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60ā88.
12. Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med 2019;25:44ā56.
13. Billah M, Waheed S, Rahman MM. An automatic gastrointestinal polyp detection system in video endoscopy using fusion of color wavelet and convolutional neural network features. Int J Biomed Imaging 2017;2017:9545920.
14. Zhang R, Zheng Y, Mak TW, et al. Automatic detection and classification of colorectal polyps by transferring low-level CNN features from nonmedical domain. IEEE J Biomed Health Inform 2017;21:41ā47.
15. Byrne MF, Chapados N, Soudan F, et al. Real-time differentiation of adenomatous and hyperplastic diminutive colorectal polyps during analysis of unaltered videos of standard colonoscopy using a deep learning model. Gut 2019;68:94ā100.
16. Repici A, Badalamenti M, Maselli R, et al. Efficacy of real-time computer-aided detection of colorectal neoplasia in a randomized trial. Gastroenterology 2020;159:512ā520.e7.
17. Komeda Y, Handa H, Matsui R, et al. Artificial intelligence-based endoscopic diagnosis of colorectal polyps using residual networks. PLoS One 2021;16:e0253585.
18. Misawa M, Kudo SE, Mori Y, et al. Artificial intelligence-assisted polyp detection for colonoscopy: initial experience. Gastroenterology 2018;154:2027ā2029.e3.
19. Wang P, Xiao X, Glissen Brown JR, et al. Development and validation of a deep-learning algorithm for the detection of polyps during colonoscopy. Nat Biomed Eng 2018;2:741ā748.
20. Itoh T, Kawahira H, Nakashima H, et al. Deep learning analyzes Helicobacter pylori infection by upper gastrointestinal endoscopy images. Endosc Int Open 2018;6:E139āE144.
21. Shichijo S, Nomura S, Aoyama K, et al. Application of convolutional neural networks in the diagnosis of Helicobacter pylori infection based on endoscopic images. EBioMedicine 2017;25:106ā111.
22. Kudo SE, Misawa M, Mori Y, et al. Artificial intelligence-assisted system improves endoscopic identification of colorectal neoplasms. Clin Gastroenterol Hepatol 2020;18:1874ā1881.e2.
23. van der Sommen F, Zinger S, Curvers WL, et al. Computer-aided detection of early neoplastic lesions in Barrettās esophagus. Endoscopy 2016;48:617ā624.
24. Horie Y, Yoshio T, Aoyama K, et al. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc 2019;89:25ā32.
25. Kanesaka T, Lee TC, Uedo N, et al. Computer-aided diagnosis for identifying and delineating early gastric cancers in magnifying narrow-band imaging. Gastrointest Endosc 2018;87:1339ā1344.
26. Wu L, Zhou W, Wan X, et al. A deep neural network improves endoscopic detection of early gastric cancer without blind spots. Endoscopy 2019;51:522ā531.
27. Zhu Y, Wang QC, Xu MD, et al. Application of convolutional neural network in the diagnosis of the invasion depth of gastric cancer based on conventional endoscopy. Gastrointest Endosc 2019;89:806ā815.
28. Ding Z, Shi H, Zhang H, et al. Gastroenterologist-level identification of small-bowel diseases and normal variants by capsule endoscopy using a deep-learning model. Gastroenterology 2019;157:1044ā1054.
29. Li P, Li Z, Gao F, et al. Convolutional neural networks for intestinal hemorrhage detection in wireless capsule endoscopy images. 2017 IEEE International Conference on Multimedia and Expo (ICME); 2017 Jul 10-14; Hong Kong, China. p. 1518ā1523.
30. Aoki T, Yamada A, Aoyama K, et al. Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc 2019;89:357ā363.e2.
31. Klang E, Barash Y, Margalit RY, et al. Deep learning algorithms for automated detection of Crohnās disease ulcers by video capsule endoscopy. Gastrointest Endosc 2020;91:606ā613.
32. Corley DA, Levin TR, Doubeni CA. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med 2014;370:2539ā2541.
33. Urban G, Tripathi P, Alkayali T, et al. Deep learning localizes and identifies polyps in real time with 96% accuracy in screening colonoscopy. Gastroenterology 2018;155:1069ā1078.e8.
34. Gong EJ, Lee JH, Jung K, et al. Characteristics of missed simultaneous gastric lesions based on double-check analysis of the endoscopic image. Clin Endosc 2017;50:261ā269.
35. Lui TK, Tsui VW, Leung WK. Accuracy of artificial intelligence-assisted detection of upper GI lesions: a systematic review and meta-analysis. Gastrointest Endosc 2020;92:821ā830.e9.
37. Kiesslich R, Burg J, Vieth M, et al. Confocal laser endoscopy for diagnosing intraepithelial neoplasias and colorectal cancer in vivo. Gastroenterology 2004;127:706ā713.
38. Mori Y, Kudo S, Ikehara N, et al. Comprehensive diagnostic ability of endocytoscopy compared with biopsy for colorectal neoplasms: a prospective randomized noninferiority trial. Endoscopy 2013;45:98ā105.
39. Kim SH, Yang DH, Kim JS. Current status of interpretation of small bowel capsule endoscopy. Clin Endosc 2018;51:329ā333.
40. Nam SJ, Lim YJ, Nam JH, et al. 3D reconstruction of small bowel lesions using stereo camera-based capsule endoscopy. Sci Rep 2020;10:6025.
41. Oh DJ, Kim KS, Lim YJ. A new active locomotion capsule endoscopy under magnetic control and automated reading program. Clin Endosc 2020;53:395ā401.
42. Milluzzo SM, Cesaro P, Grazioli LM, et al. Artificial Intelligence in lower gastrointestinal endoscopy: the current status and future perspective. Clin Endosc 2021;54:329ā339.
43. Stidham RW, Liu W, Bishu S, et al. Performance of a deep learning model vs human reviewers in grading endoscopic disease severity of patients with ulcerative colitis. JAMA Netw Open 2019;2:e193963.
44. Maeda Y, Kudo SE, Mori Y, et al. Fully automated diagnostic system with artificial intelligence using endocytoscopy to identify the presence of histologic inflammation associated with ulcerative colitis (with video). Gastrointest Endosc 2019;89:408ā415.
45. Ozawa T, Ishihara S, Fujishiro M, et al. Novel computer-assisted diagnosis system for endoscopic disease activity in patients with ulcerative colitis. Gastrointest Endosc 2019;89:416ā421.e1.
46. Liu Y, Chen PC, Krause J, et al. How to read articles that use machine learning: usersā guides to the medical literature. JAMA 2019;322:1806ā1816.
47. Eelbode T, Sinonquel P, Maes F, et al. Pitfalls in training and validation of deep learning systems. Best Pract Res Clin Gastroenterol 2021;52-53:101712.
48. der Pol CBV, Tang A. Imaging database preparation for machine learning. Can Assoc Radiol J 2021;72:9ā10.
49. Sutton RA, Sharma P. Overcoming barriers to implementation of artificial intelligence in gastroenterology. Best Pract Res Clin Gastroenterol 2021;52-53:101732.
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
